Mastermind Release Notes: See the Latest Updates
This is a running account of enhancements made to the Mastermind Genomic Search Engine. The most recent updates are at the top, followed by previous updates, so you can see all that the Genomenon team is doing to improve Mastermind in one place.
Enhancing the Mastermind User Experience Through Streamlined Workflows & Expanded Content
The team at Genomenon continues to build on its product innovations with the latest release of Mastermind, designed to further help diagnostic labs speed up workflows and quickly assess evidence for patient variants.
Mastermind Release Notes: Mastermind 3.0
Over the last year, the dedicated team at Genomenon have been working tirelessly to understand what users want and need from Mastermind. Today, we are launching the first step toward an enhanced user experience and user interface with the release of Mastermind’s new Gene Information Page.
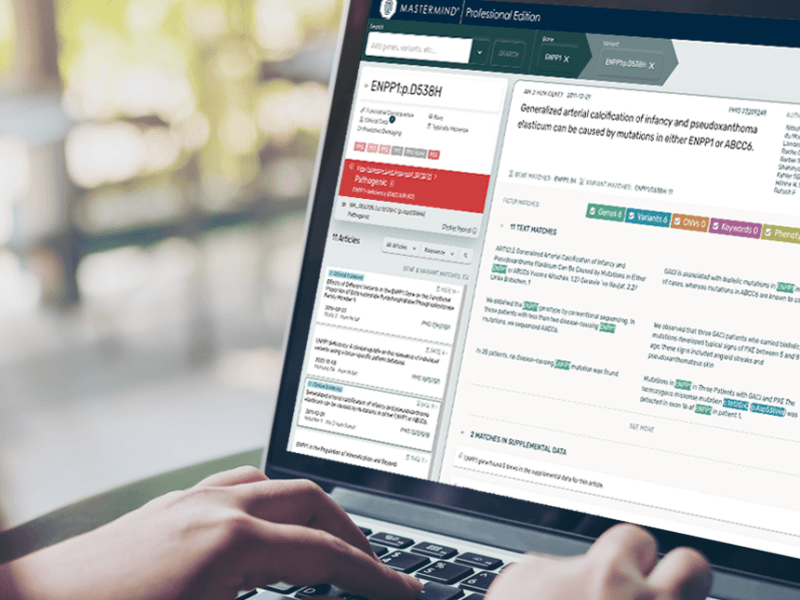
Mastermind Update: Finding Your Supplemental Match
The Mastermind team has released its latest update to the Genomic Search Engine platform, which aims to help users find their supplemental match and filter by PMID. This update is a significant development for Mastermind, as it takes into account user feedback to provide more personalized and accurate search results.
Mastermind User Group Recap – Salt Lake City, Spring 2023
Last week, the Genomenon team hosted its inaugural User Group Meeting in Salt Lake City. For us, this highly anticipated gathering of Mastermind users was truly special as it marked the beginning of an exciting new tradition of learning and collaboration.
ClinVar in Mastermind
The latest update to Mastermind has integrated ClinVar, a public database of clinically observed variants and their pathogenicity interpretations by the clinical labs that submitted the variants. This can often serve as a quick jumping off point for deciding to further investigate a given variant based on the evidence provided in Mastermind.