Fusion genes have long been known to play an important role in the development of cancer. Identifying and documenting each newly discovered fusion is crucial in both patient diagnosis and the development of Precision Medicine.
The Genomenon team used the Mastermind Genomic Search Engine to compile a comprehensive knowledgebase of literature regarding gene fusions in order to analyze the data and relate the findings in this report.
Fusion Genes: An Emerging Target in Cancer Diagnostics and Drug Development
Fusion genes have long been known to play an important role in the development of cancer.1 Since the discovery of the first fusion gene in 1960, researchers have continued to discover fusion events involved in cancer development.
In recent years, hundreds of novel fusions have been identified across a multitude of cancer types, due in large part to the ease of producing this data using next generation sequencing (NGS) techniques like RNAseq and their broad use in clinical diagnostic labs. The progression of gene fusions from discovery to characterization to drug development represents a new and increasingly common occurrence.
Identifying and documenting each newly discovered fusion is crucial in both patient diagnosis and the development of precision medicine. This diagnostic method allows for the proper application of existing therapies and the development of new therapies.
It came to our attention that a comprehensive knowledgebase of literature regarding gene fusions was not available, so we set out to create one from the Mastermind genomic database. Below are the results of that effort.
A Brief History of Fusion Gene Discoveries
The first known fusion gene, BCR-ABL1, was discovered initially as an aberrantly small chromosome based on cytogenetic analysis of Chronic Myelogenous Leukemia (CML) cells in 1960. It was later characterized in 1973 as an abnormal translocation of chromosomes 9 and 22. Then, in the 1980s, it was understood to cause the fusion of two gene products – BCR and ABL1. This fusion gene is now known to be present in more than 95% of CML patients and 5-25% of Acute Lymphoblastic Leukemia patients.2-4 Imatinib, used for the treatment of BCR-ABL1 positive CML5, was among the first therapeutic compounds to be developed to specifically target a genetic lesion – 23 years after the first discovery of a fusion gene.
A 2017 study reported that 15% of patients with metastatic cancer harbored genomic rearrangements, many of which produced putative fusions.6 While 35% of these fusions involved kinase genes, indicating they may able to be targeted by currently existing kinase inhibitors, 19% of them involved novel partner genes. This discovery left open the possibility for new strategies of drug development.6
In a large-scale 2018 study, a total of 25,000 fusions were discovered across 9,600 tumor samples encompassing 33 different cancer types.7 Fusion events drove pathogenesis of 16.5% of these cancers, and were the sole driver in 1.8% of them.7 Additionally, 6% of the samples contained fusions that could potentially be targeted by currently existing therapies.7
Fusion Gene Knowledgebases
Currently, The Catalogue of Somatic Mutations in Cancer (COSMIC) and The Cancer Genome Atlas (TCGA) serve as the main source for documented fusions.8-9 COSMIC currently contains 297 unique fusion pairs derived from ~ 1.4 million tumor samples, while TCGA contains clinical and sequencing data from over 20,000 samples.8, 10-11
The comprehensiveness of these databases is of the utmost importance due to these factors:
- Many fusions discovered in tumor samples involve novel partner genes
- These fusions can often be targeted by existing or emerging therapies
However, both COSMIC and TCGA lack the comprehensive literature support needed to ensure that the database is fully inclusive of all documented fusions. Moreover, these fusions being aggregated from large patient sequencing studies without more detailed curation introduces the potential for the inclusion of incidental fusion events that do not drive disease, non-functional read-through fusions, or sequencing and bioinformatic artifacts.
To address this need to improve clinical diagnosis for fusion events and to inform drug development in pharmaceutical workflows for gene fusion-based therapeutics, we used the Mastermind Genomic Search Engine to produce a comprehensive landscape of all previously published fusion events, and to characterize their role in disease.
Mastermind: A More Comprehensive Source for Fusion Documentation
The Mastermind Genomic Database has indexed the full text of millions of genomic articles and supplemental data to provide immediate insight into the published research for every disease, gene, and variant found in the literature. Access to this annotated information allowed us to obtain more complete answers to questions about the comprehensive fusion landscape in human cancers.
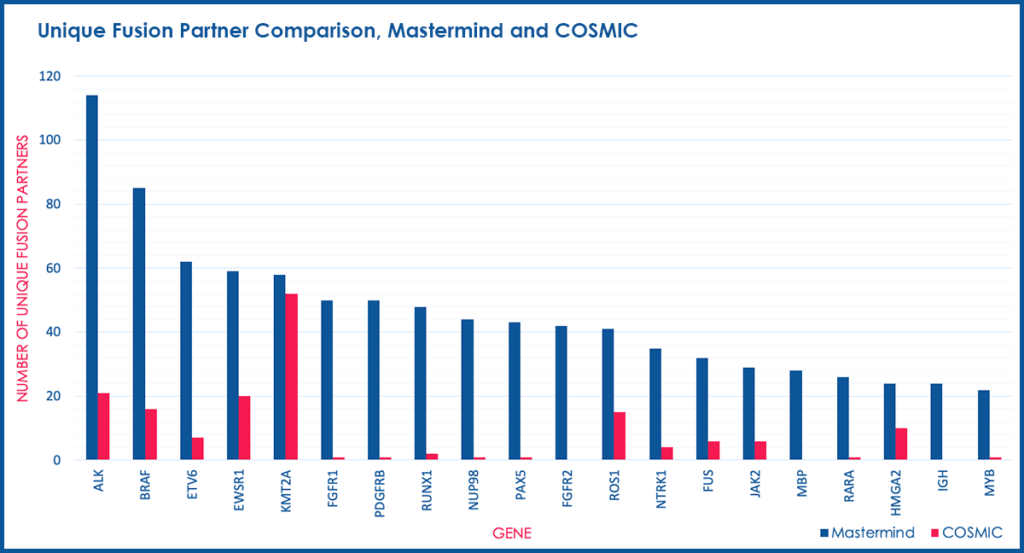
In order to determine whether Mastermind could serve as a more comprehensive source for documented fusions, we developed a process to automatically retrieve fusion genes from our database of approximately 6.5 million full-text genomic articles. To focus our study on fusion events of clinical significance, we restricted our analysis to the 507 genes comprising the Illumina TruSight Fusion Gene Panel. For these 507 genes, we discovered a total of 2,022 unique fusion pairs cited in the scientific literature, all of which were manually validated. This represents a 686% increase in yield over the COSMIC database (Figure 1).
Figure 1. Unique Fusion Partner Comparison, Mastermind and COSMIC
NPM1-TYK2 is an example of a fusion gene that is not present in either the COSMIC or the TCGA databases, but for which there is a known pathogenic disease association and a potential therapy. This fusion was discovered in 2014 and found to be recurrent in cutaneous CD30-positive lymphoproliferative disorders.12 Because the fusion event involves activation of the JAK-STAT pathway member TYK2, it represents a potentially targetable fusion event.
Top Fusion Partners by Number of Articles
The top 5 most common fusion partners:
- ALK (n = 94 unique fusion partners)
- BRAF (n = 85 unique fusion partners)
- ETV6 (n = 62 unique fusion partners)
- EWSR1 (n = 59 unique fusion partners)
- FGFR1 (n = 50 unique fusion partners)
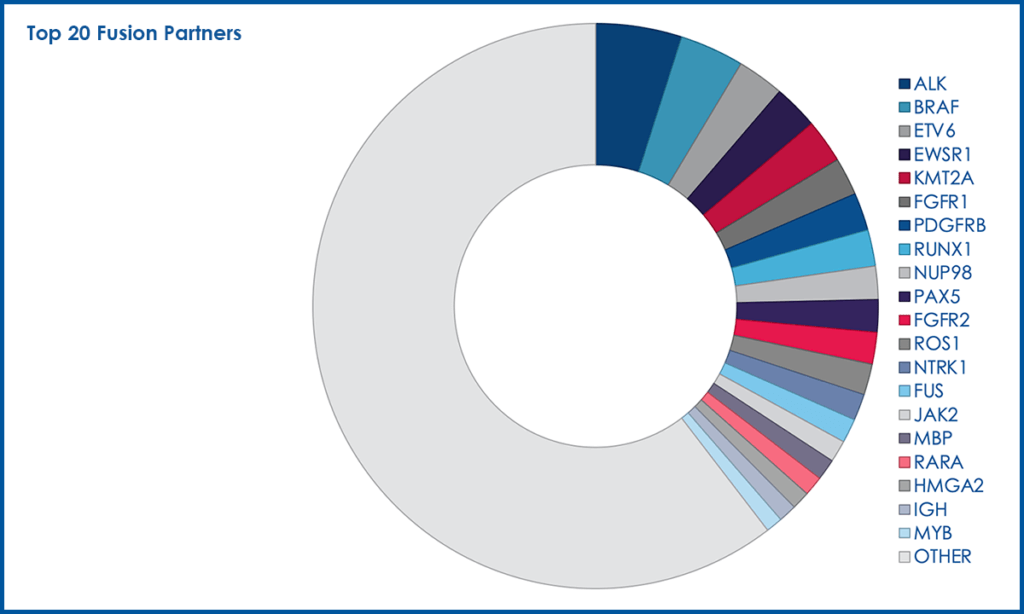
Overall, the top 20 most common fusion partners represented 22.6% of the total fusion partners (Figure 2).
Figure 2. Top 20 Fusion Partners
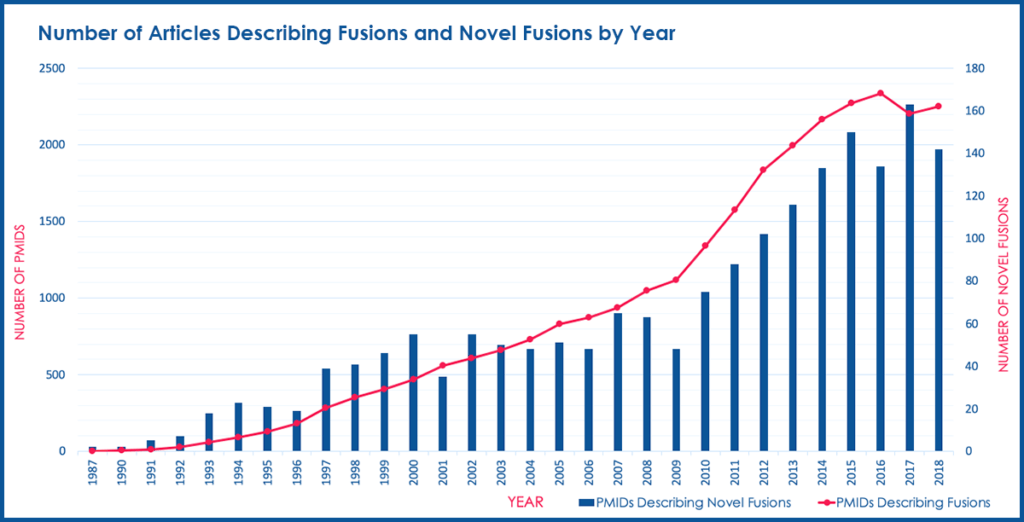
Based on the publication date of the articles describing these fusions, we also discovered that both the total number of articles describing fusions and the number of articles describing novel fusions has experienced a relatively constant increase from 1987 to 2018 (Figure 3).
Figure 3. Number of Articles Describing Fusions and Novel Fusions by Year
This trend represents a steady increase in recognition of fusion genes in clinical diagnostic labs and research laboratories. Last year over 140 novel gene fusions were found across nearly 500,000 newly published scientific articles containing genomic content. This highlights the challenges associated with maintaining a current and comprehensive database of fusion events that can lead to disease and how to most effectively treat patients who have these fusion events.
New Fusion Genes for Cancer in 2019
From among the newly published scientific articles in 2019 alone, we have used Mastermind to identify 11 novel fusions involved in the genesis of multiple cancer types (Table 1).
| Fusion | PMID | Title | Cancer Type |
|---|---|---|---|
| AUTS2-KMT2C | 30718424 | Genome-Wide Colocalization of RNA-DNA Interactions and Fusion RNA Pairs | Lung Cancer |
| BRAF-SEPT3 | 30254212 | BRAF Fusions Identified in Melanomas Have Variable Treatment Responses and Phenotypes | Melanoma |
| ETV6-IKZF1 | 30643249 | PAX5-Driven Subtypes of B-Progenitor Acute Lymphoblastic Leukemia | B-Progenitor Acute Lymphoblastic Leukemia |
| LPP-RFC4 | 30649385 | The Genomic Landscape of Mucinous Breast Cancer | Mucinous Breast Cancer |
| NTRK3-KHDRBS1 | 30187166 | Novel KHDRBS1-NTRK3 Rearrangement in a Congenital Pediatric CD34-Positive Skin Tumor: a Case Report | Congenital CD34-Positive Dermohypodermal Spindle-Cell Neoplasm |
| PDGFRB-GCC2 | 30697976 | A Novel Fusion Gene Involving PDGFRB and GCC2 in a Chronic Eosinophilic Leukemia Patient Harboring t(2;5)(q37;q31) | Chronic Eosinophilic Leukemia |
| ROS1-KLC1 | 30350109 | Identification of a Novel KLC1-ROS1 Fusion in a Case of Pediatric Low-Grade Localized Glioma | Pediatric Low-Grade Localized Glioma |
| EWSR1-SMAD3 | 30709442 | Mesenchymal Tumors with EWSR1 Gene Rearrangements | Mesenchymal Neoplasms |
| ALK-CAMKMT | 30579547 | A Novel CAMKMT Exon 3-ALK Exon 20 Fusion Variant was Identified in a Primary Pulmonary Mucinous Adenocarcinoma | Primary Pulmonary Mucinous Adenocarcinoma |
| CBFA2T3-PAX5 | 30643249 | PAX5-Driven Subtypes of B-Progenitor Acute Lymphoblastic Leukemia | B-Progenitor Acute Lymphoblastic Leukemia |
| ROS1-COL4A3BP | 30719217 | ROS1-GOPC/FIG: A Novel Gene Fusion in Hepatic Angiosarcoma | Synovial Sarcoma |
| ROS1-NETO1 | 30719217 | ROS1-GOPC/FIG: A Novel Gene Fusion in Hepatic Angiosarcoma | Perivascular Epithelioid Cell Tumor |
Table 1. Novel Fusions Discovered in 2019
Several of these fusions were unique, having been discovered in an individual patient:
- BRAF-SEPT3 is a novel fusion discovered in one patient with melanoma and conferred the least proliferative but most invasive phenotype of the three BRAF fusions that were evaluated, as well as a low treatment response to MAPK inhibitors.13
- Similarly, NTRK3-KHDRBS1 was discovered in an infant with a CD34-positive spindle-cell skin tumor and is of particular interest due to the more recent identification of the role of NTRK3 fusions in driving the development of rare cancer types, as well as the recent development of TRK inhibitors.14
Other fusions were detected in more than one case:
- LPP-RFC4, is a recurrent but non-pathogenic fusion in mucinous breast cancer15
- CBFA2T3-PAX5 is a recurrent fusion in a high-risk subtype of B-progenitor acute lymphoblastic leukemia16
Overall, these findings illustrate an increased interest in the role of fusions in cancer generally, as well as emphasize the true breadth and heterogeneity of the fusion landscape.
Summary of Findings
As our knowledge of gene fusions and their function in the development of cancer continues to improve, identifying and documenting each newly discovered fusion will only become more crucial in both patient diagnosis and the development of precision medicine. This will allow for application of existing therapies and the development of new therapies as fusions are more thoroughly characterized and novel fusions are discovered.
A comprehensive view of the fusion landscape in cancer can be developed by indexing the entire corpus of the genetic literature for gene fusions. The resulting gene fusion database can provide insight into patient diagnosis and treatment decisions, and provide a platform for both drug discovery and repurposing efforts.
Request access to the complete Mastermind Gene Fusion Database here.
The content as presented in this article is an analysis of the literature on fusion genes as of April 2019, and provided by Genomenon team members Lauren Chunn, Mark Kiel, and Diane Nefcy.
Mastermind, Genomenon’s Genomic Search Engine, provides immediate insight into the published genomic research for every disease, gene, and genetic variant found in the literature.
Used by hundreds of diagnostic labs around the world, Mastermind accelerates genomic interpretation by providing unique insight into genomic relationships found in the full text of millions of scientific articles.
Pharmaceutical researchers license the Mastermind database for a comprehensive genomic landscape associated with any given disease – to identify and prioritize genomic biomarkers for drug discovery and clinical trial targets.
References
- Mitelman, Felix, Johansson, Bertil, and Mertens, Fredrik. “The impact of translocations and gene fusions on cancer causation.” Nature Reviews Cancer vol. 7,4 (2007): 233-245.
- Parker, Brittany C., and Zhang, Wei. “Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment.” Chinese journal of cancer vol. 32,11 (2013): 594-603.
- Orna, Dreazen et al. “Multiple molecular abnormalities in Ph1 chromosome positive acute lymphoblastic leukaemia.” British Journal of Haematology vol. 67,11 (1987): 319-324.
- Nowell, P. C. and Hungerford, D. A. “A minute chromosome in human chronic granulocytic leukemia.” Science 132, (1960): 1497.
- Buchdunger, Elizabeth et al. “Inhibition of the Abl Protein-Tyrosine Kinase in Vitro and in Vivo by a 2-Phenylaminopyrimidine Derivative.” Cancer Research vol. 56,1 (1996): 100-104.
- Zehir, Ahmet et al. “Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients.” Nature medicine vol. 23,6 (2017): 703-713.
- Gao, Qingsong et al. “Driver Fusions and Their Implications in the Development and Treatment of Human Cancers.” Cell reports vol. 23,1 (2018): 227-238.
- Tate, John G. et al. “COSMIC: the Catalogue Of Somatic Mutations In Cancer.” Nucleic acids research vol. 47,D1 (2018): D941-D947.
- Tomczak, Katarzyna et al. “The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge.” Contemporary oncology vol. 19,1A (2015): A68-A77.
- https://cancer.sanger.ac.uk/cosmic/fusion
- https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
- Velusamy, Thirunavukkarasu et al. “A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders.” Blood vol. 124, 25 (2014): 3768-71.
- Turner, Jacqueline A. “BRAF fusions identified in melanomas have variable treatment responses and phenotypes.” Oncogene vol. 38,8 (2019): 1296-1308.
- Tallegas, Matthias et al. “Novel KHDRBS1-NTRK3 rearrangement in a congenital pediatric CD34-positive skin tumor: a case report.” Virchows Archiv vol. 474, 1 (2019): 111-115.
- Pareja, Fresia et al. “The Genomic Landscape of Mucinous Breast Cancer.” Journal of the National Cancer Institute (2019). [Epub ahead of print]
- Gu et al. “PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia” Nature Genetics 51 (2019): 296-307.