The UCSC Genome Browser provides free, public access to genome assemblies, allowing researchers and clinicians the opportunity to search the genomic data of humans and many other species. Academics and researchers have been utilizing this interactive suite of tools, evidence, and downloadable data files for nearly 20 years.
Mastermind has been a part of the UCSC Genome Browser for some time, and has recently added Mastermind to its GRCh38/hg38 human assembly.
Here’s how to add the Mastermind track to the UCSC Browser and link to Mastermind results.
Step 1.
Start from UCSC Genome Browser Homepage and click “Genome Browser” link under “Our tools”: http://www.genome.ucsc.edu/
Step 2.
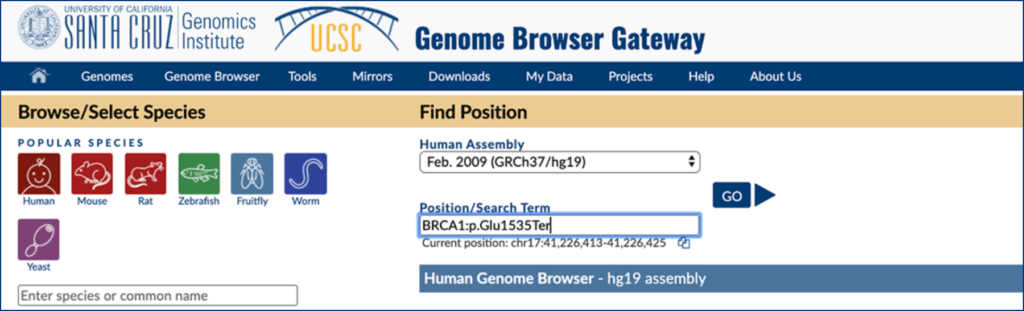
Choose “GRCh37/hg19” or “GRCh38/hg38” option under “Human Assembly”, and search for your variant (Proper capitalization and format is necessary)
Step 3.
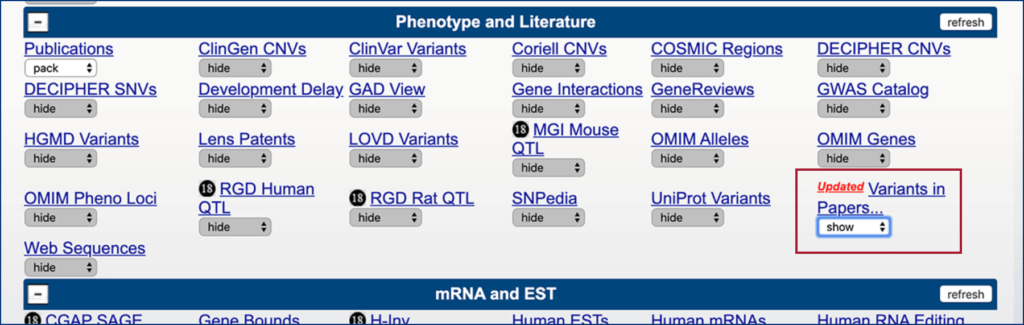
Scroll down to “Phenotype and Literature” section and select “Show” for “Variants in Papers…” and then click “Refresh” button above next to “Phenotype and Literature”:
Step 4.
Scroll up to genome browser at the top of page, right click on the condensed “Genomenon Mastermind…” track, and click “full”. If you receive an error message, zoom in closer.
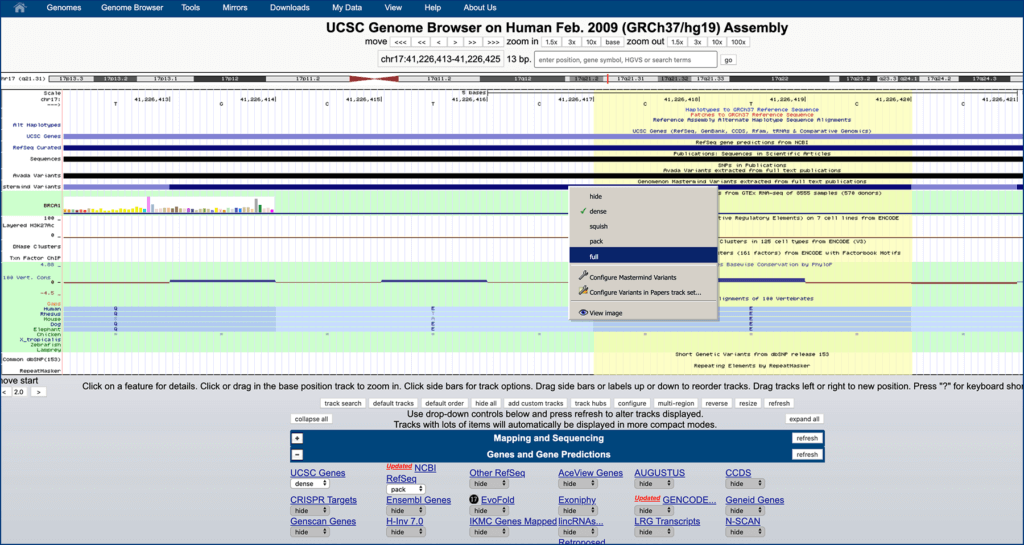
Step 5.
Find your variant in the highlighted region (from your search in step 2), and click the desired variant entry in the “Genomenon Mastermind” track to access the detail page.
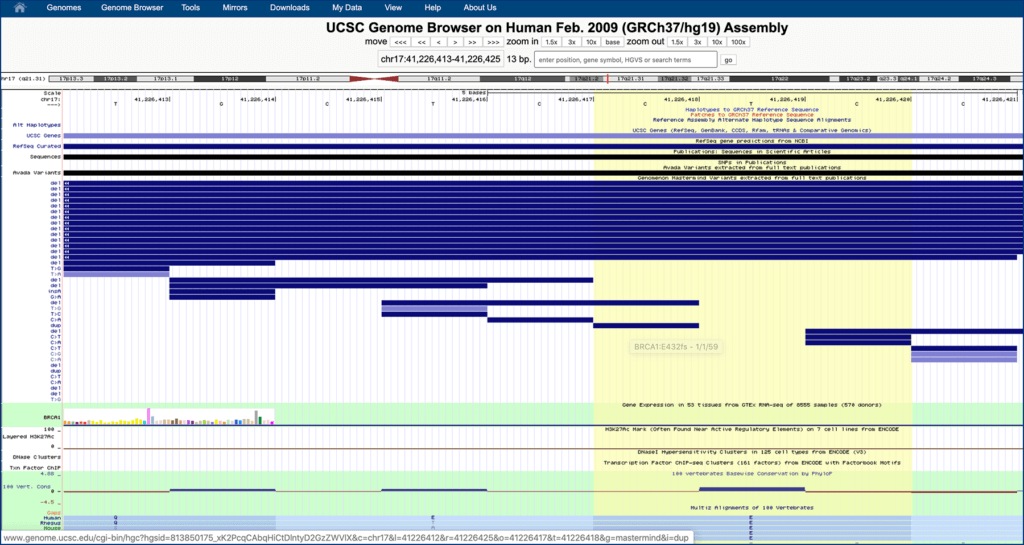
Step 6.
Click the links in the “Protein change and link to details” box to view articles in Mastermind.
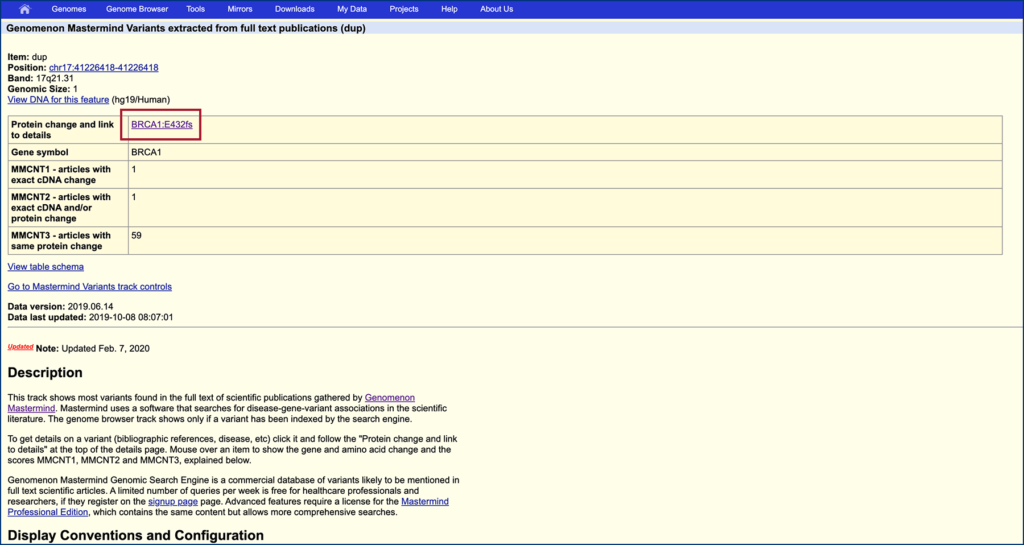
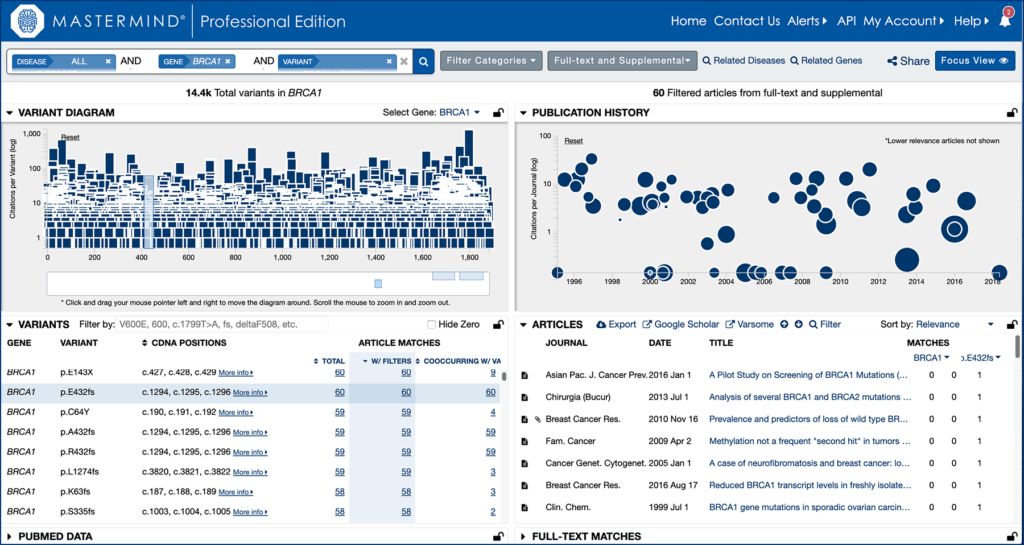
Step 7.
Explore evidence related to your variant in Mastermind.
Questions about Mastermind or this blog? Email us
To learn more about our integration with the UCSC Genome Browser, read the press release.