The Mastermind Genomic Search Engine now includes searching by Copy Number Variation (CNV). This new feature accelerates the diagnosis and interpretation of results for patients with genetic disease. Here’s how to find CNVs in Mastermind.
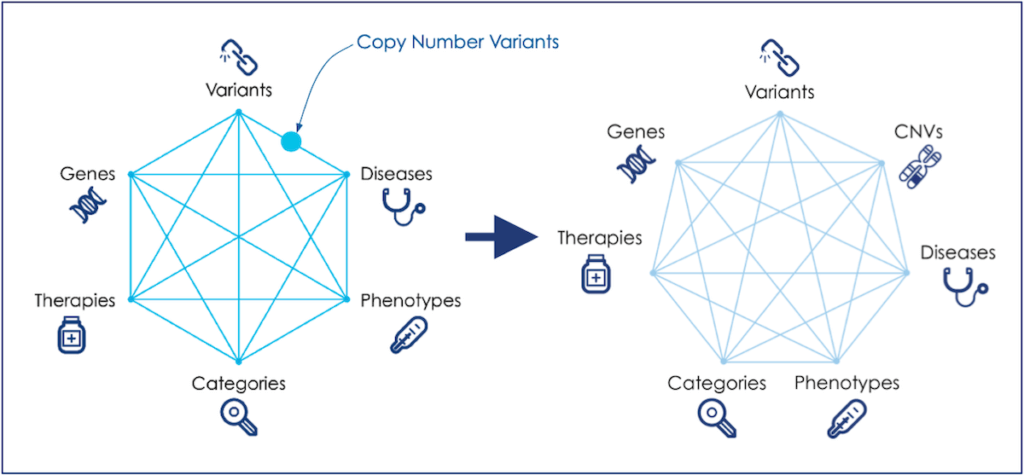
Mastermind identifies every gene, variant, disease, phenotype, therapy, and categorical keyword cited in the evidence of the medical literature, in every way that an author can describe them. Mastermind’s Genomic Association Intelligence then normalizes these various descriptions (or nomenclature) of each concept, and brings to light the associations linking any two or more entities with the supporting evidence.
This knowledge allows users to know, for example, which variants are associated with which disease, which genes and diseases are associated with which therapies, etc. The use-cases enabled by identifying these associations have now grown considerably with the addition of CNVs.
The Mastermind “Association Intelligence” hexagon is now a septagon!
The primary difficulties with CNV data are twofold:
- the heterogeneity of author descriptions across the medical literature.
- the evaluation for the relevance of the effectual part of a CNV related to some disease.
How to Describe CNVs
There are many ways in which to describe a structural change in DNA as a CNV. Common CNV descriptions include:
- By cytogenetic band
E.g. “Duplication of chromosome band 11q23” - ISCN karyotype notation
E.g. “46,XX,del(5)(q13)” or “dup(1)(p36p11)” - Genomic coordinates
E.g. “deletion (chr11:1918222-1977026 (hg18))” - Genes and exons
E.g. “duplication of MLL exons 2-6”
Mastermind’s Genomic Language Processing capabilities recognize these nomenclatures, among others, and normalize these descriptions to genomic coordinates with a start position, end position, and effect type (deletion or duplication/amplification). This allows users to search for CNVs using any of these nomenclatures, and returning all relevant citations regardless of the nomenclature used in each citation.
To search for a CNV in Mastermind, enter the desired CNV into the search input using one of the nomenclatures above. Mastermind uses Natural Language Processing (NLP) to recognize and understand the description you enter for the CNV. You will know if Mastermind recognizes the CNV search when it displays a suggestion with a “CNV” label:
At minimum, the entered description should have some indication of the location and the effect type (deletion or duplication/amplification).
The Size of CNVs
Whereas a variant is most often associated with a single gene, which codes for some protein (or proteins) and affects some biological process, CNVs can span hundreds or thousands of genes, making it difficult to narrow down to the causative region of interest.
Additionally, with such large structural changes, there are diminishing chances of the searched CNV having been cited in the literature precisely at the same arbitrary start and end position on a chromosome (this isn’t always true in practice due to CNVs commonly being studied and described at convenient breakpoints, such as deletions or amplifications of a whole gene or specific exons within a gene, but it’s true in general).
How to find CNVs
Articles List
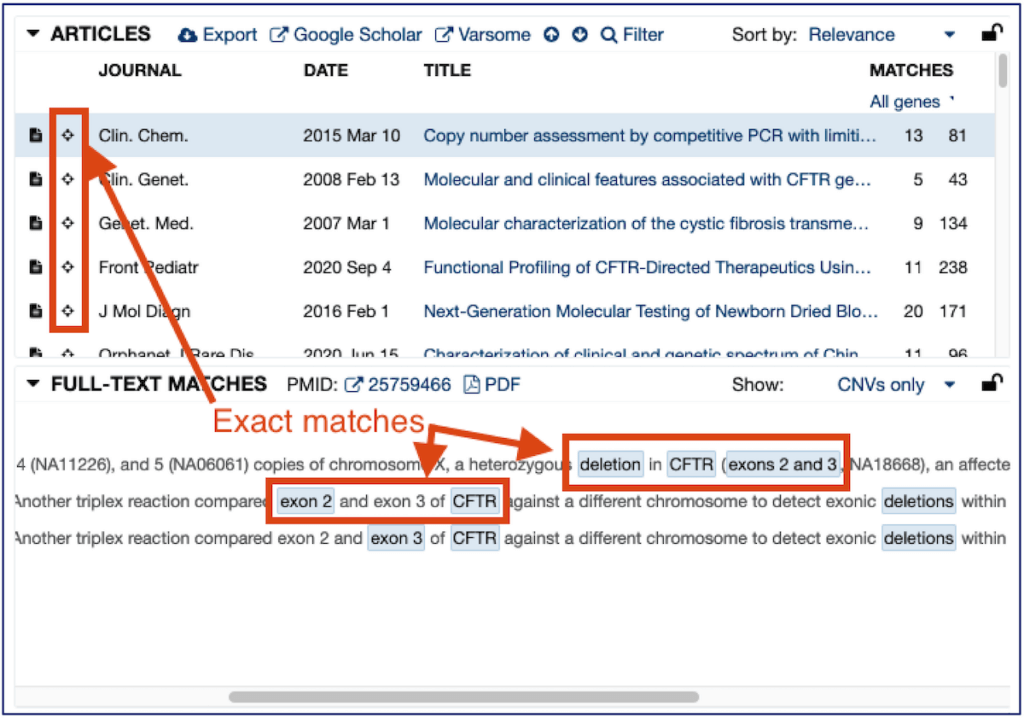
For the reasons above, the CNV search functionality in Mastermind functions differently than the variant search. While the variant search shows the results for the exact variant being searched, the CNV search will show results citing any overlapping CNV to that being searched by default. If there are any exactly-matched CNVs in the literature, those results will be prioritized at the top of the results, with a “bullseye” icon.
If you wish to filter results to only those which match the searched CNV exactly, a filter is provided at the top, which allows toggling between “Overlapping CNVs” and “Exact CNV”:

CNVs List
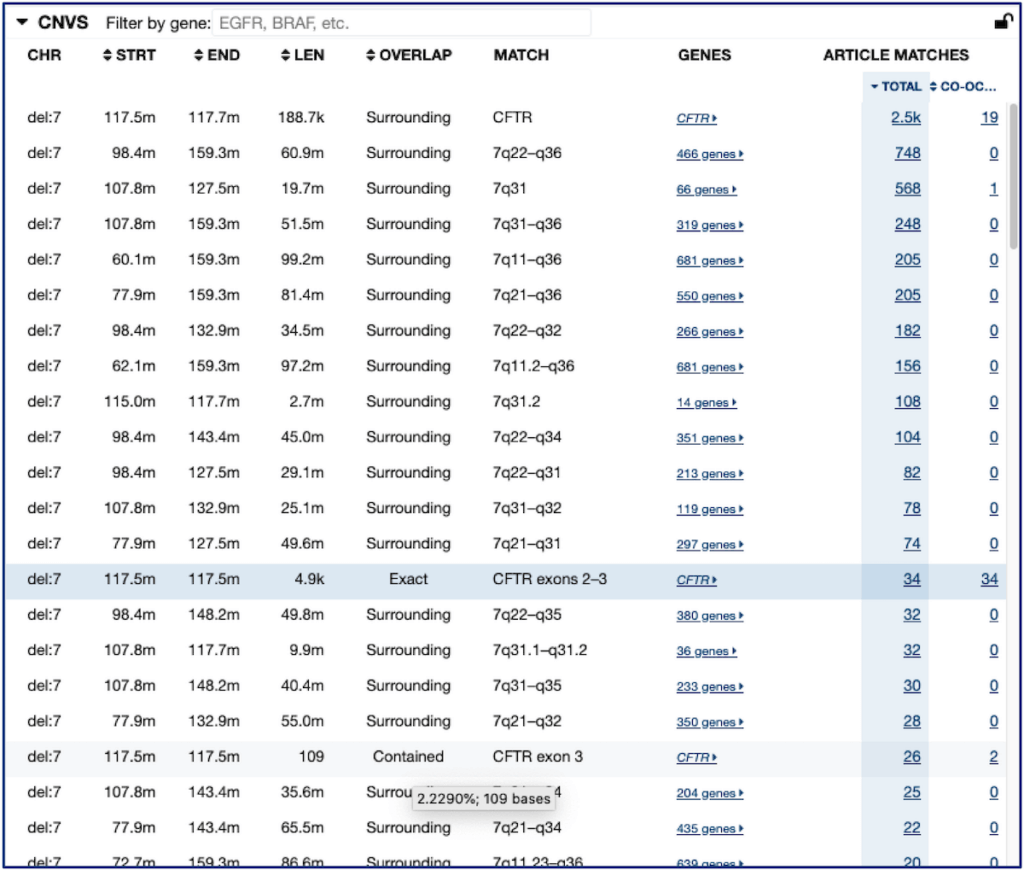
You will also notice that in the left panel of the “Detail” page showing the search results, all of the overlapping CNVs being searched will be shown instead of a list of variants, as is shown for variant searches in Mastermind.
The columns shown for the CNV list are:
- CHR – the effect type (“del” for deletion, “amp” for duplication or amplification) and chromosome number.
- STRT – the genomic coordinate (using GRCh38) of the start position on the chromosome.
- END – the genomic coordinate (using GRCh38) of the end position on the chromosome.
- LEN – the length of the CNV in nucleotide bases.
- OVERLAP – the type of overlap between each CNV in the list and the searched CNV.
– “Surrounding” means the listed CNV is larger and completely engulfs the searched CNV.
– “Intersecting” means the listed CNV partially overlaps the searched CNV
– “Contained” means the listed CNV is smaller than and contained within the searched CNV.
– “Exact” means the listed CNV is the searched CNV.
– In addition to the listed overlap type, you can hover the cursor over the above values to view the reciprocal overlap calculation between the searched CNV and each CNV in the list. For example, the exact CNV will be 100%, and other CNVs will be the mutual overlap between the two CNVs, shown both as a percentage and as the number of overlapping bases. When you sort by this column, it sorts by the reciprocal overlap percentage. - MATCH – the best match description of the CNV, making the CNV easier to understand than trying to remember start and end coordinates.
- GENES – the genes which the listed CNV overlaps.
- ARTICLE MATCHES – the number of articles exactly matching the listed CNV.
The CNV list is sorted by number of articles citing that CNV, by default. Other headers may be clicked to sort by those instead.
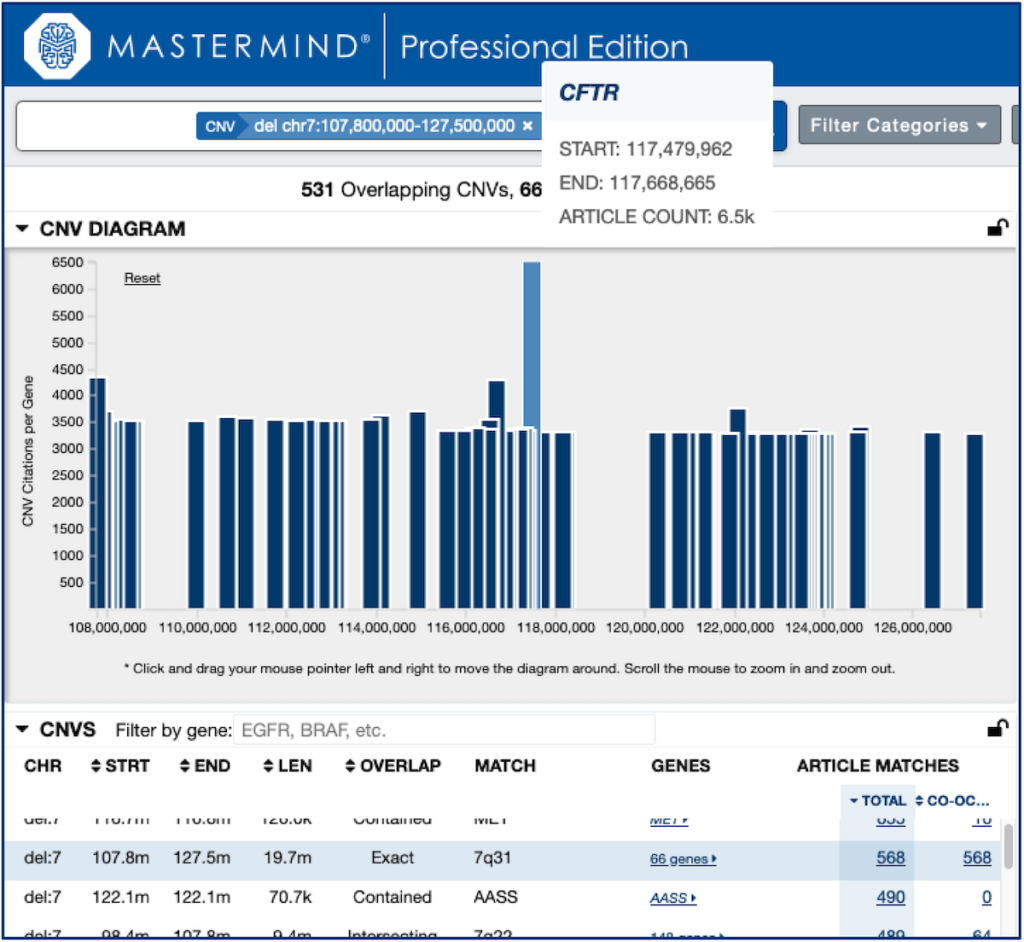
CNV Diagram
The CNV diagram shows the gene hotspots for the searched CNV, making it easy to find the gene[s] likely to be relevant to the biological impact of the CNV. The genes are plotted by their start positions on the chromosome along the X-axis, and the number of CNV citations overlapping that gene on the Y-axis. Using the diagram makes it easy to zoom in and hover over the hotspots to see which genes they are, and then clicking the gene will add it to the search, further filtering the results to the literature which explicitly discusses those genes in the context of the searched CNV.
Try Mastermind for yourself today or talk to us about upgrading to Pro to use the new CNV search feature.