DNASTAR’s Variant Annotation Database (VAD), included in Lasergene Genomics, is now integrated with the Mastermind Genomic Search Engine. With this integration, Lasergene users can now quickly and easily search and cross-reference NGS variant data from millions of PubMed publications.
DNASTAR Lasergene software supports molecular biologists, geneticists, and clinical researchers by meeting virtually all of their DNA, RNA, and protein analysis needs, including Sanger and next-generation sequence (NGS) assembly and analysis, variant detection and analysis, protein sequence and structure analysis, and protein modeling. The Lasergene Genomics package enables researchers to set up complex genomic sequencing projects in mere minutes and automates tasks that typically require extensive manual intervention with other software packages. Integrated analysis allows you to see and understand your sequencing results with ease.
Here is how you can view Mastermind results from the DNASTAR Lasergene Genomics platform:
STEP 1.
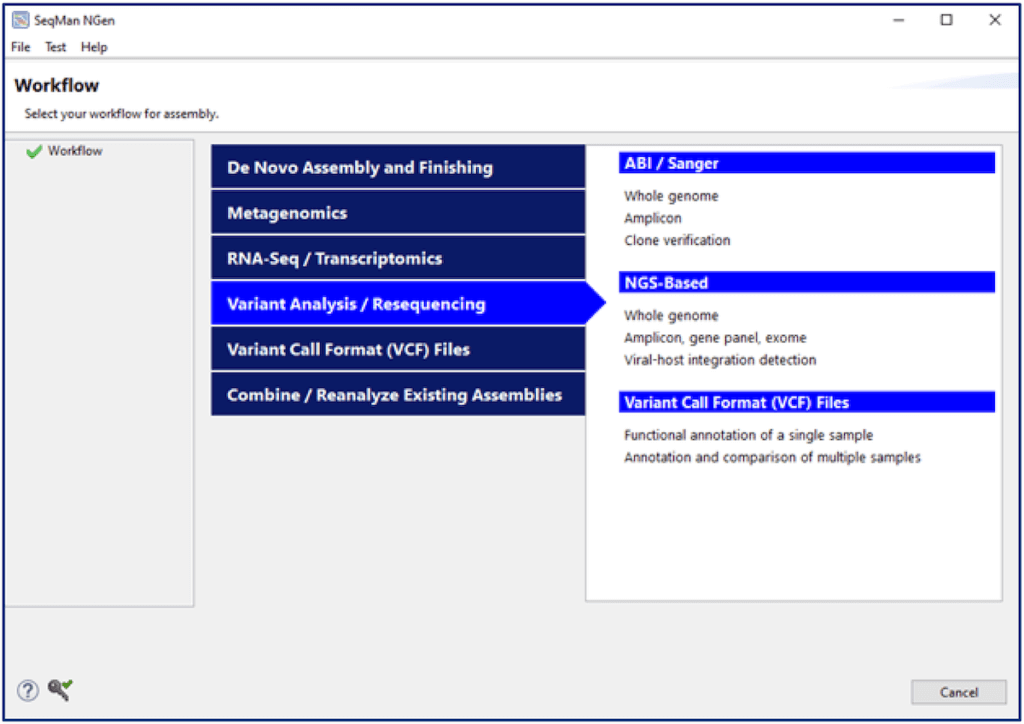
Launch the SeqMan NGen application installed with Lasergene and choose the appropriate workflow for your input data from the Variant Analysis/Resequencing tab. Note that you can import Sanger or NGS reads for assembly or use the Variant Call Format (VCF) File workflow if your data has already been assembled and the variants detected and saved to VCF format. SeqMan NGen will guide you through the setup for one or multiple samples so that read assembly (or VCF import), variant detection, variant filtering, sample cross-comparison and variant annotation are all run simultaneously and automatically.
STEP 2.
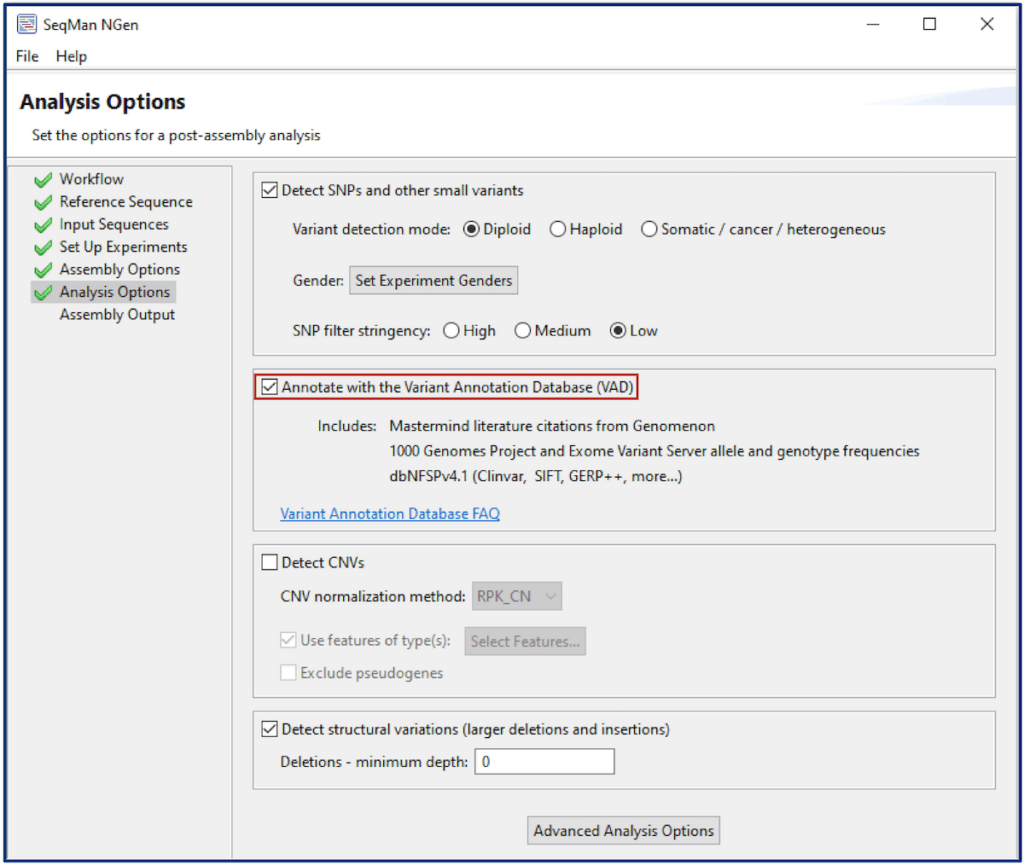
Proceed through your project setup following the on-screen prompts in SeqMan NGen. On the Analysis Options page, choose the option to Annotate with the Variant Annotation Database (VAD). Choosing this option will automatically annotate the variants detected in all the samples in your project from multiple databases, including Mastermind citation counts and hyperlinks to the Mastermind Genomic Search Engine.
STEP 3.
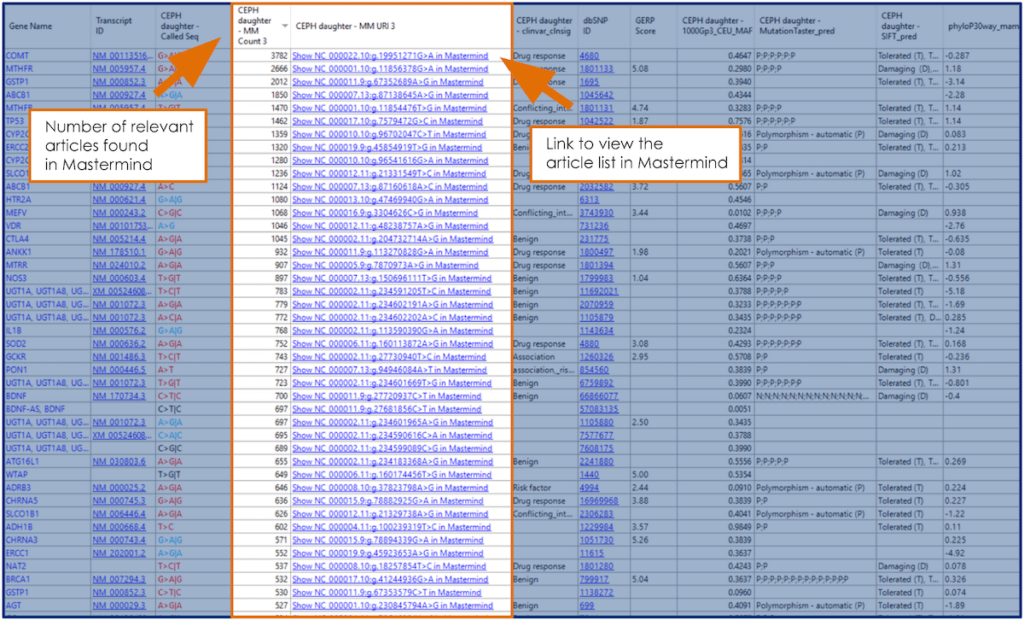
Continue through your project setup and run the assembly following the prompts in SeqMan NGen. Then open the project in ArrayStar to analyze variants. The variant table in ArrayStar is automatically populated with Mastermind counts (MM Count 3) and hyperlinks (MM URI 3) as well as other annotations including (pictured): GERP, ClinVar, 1000 Genomes minor allele frequencies, Mutation Taster predictions, and SIFT predictions. Click on links in the MM URI 3 column for your variants of interest to view, filter, and analyze the article list in Mastermind.
STEP 4.
When viewing the evidence in Mastermind, you can take your search further with the Filter Categories feature. Filter and analyze your chosen criteria by ACMG interpretation, clinical significance, genetic mechanism, and significant terms in the abstract.
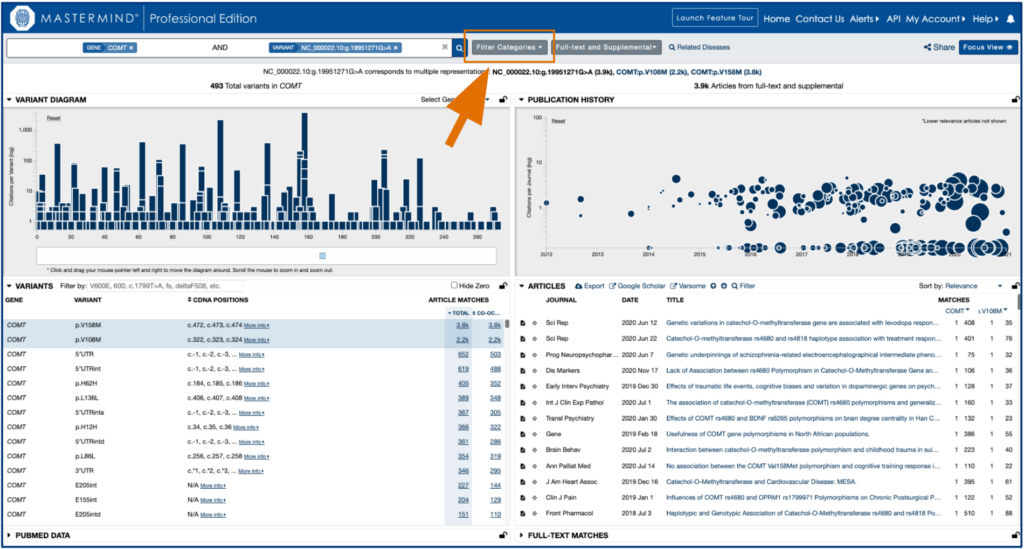
STEP 5.
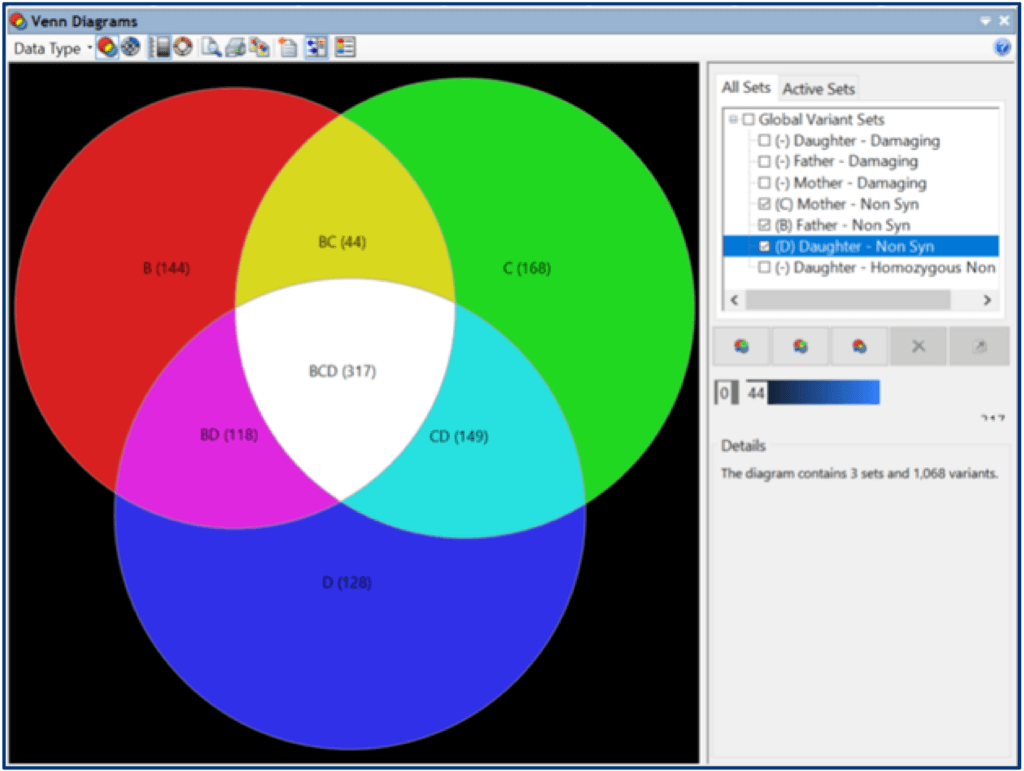
Within ArrayStar, you can filter variants based on the Mastermind and other annotation databases, as well as SNP functional classification. Create sets to save genes and SNPs of interest. Sets can be visualized and compared in Venn diagrams and other graphical views.
Detailed tutorials for variant analysis in Lasergene can be found here.
See all of the Trusted Variant Interpretation Solutions that Mastermind is integrated with.
Try Mastermind for yourself. Start your free trial of Professional Edition today.