Mastermind now has greatly enhanced capabilities when searching for intronic and non-coding variants across the medical literature. I’m covering this major enhancement in two blog posts. In this post, I’ll demonstrate how we’ve applied this approach to improve the precision and prioritization of non-coding variant (and other nucleotide-specific variant nomenclature) searches in Mastermind.
In my last post, Sensitivity versus Specificity for Non-Coding Variants, we discussed the differences between nucleotide-specific and codon-specific variant descriptions, and the trade-offs between sensitivity and specificity, especially for non-coding variants.
We’ve refined our groupings of non-coding variants to be more specific, and prioritized results that do match the nucleotide-specific description searched at the top of the search results. This helps ensure the most relevant and important results for the search aren’t missed, while avoiding the trade-off with sensitivity for articles which still appear further down in the results.
Prioritized Results for Nucleotide-Specific Searches
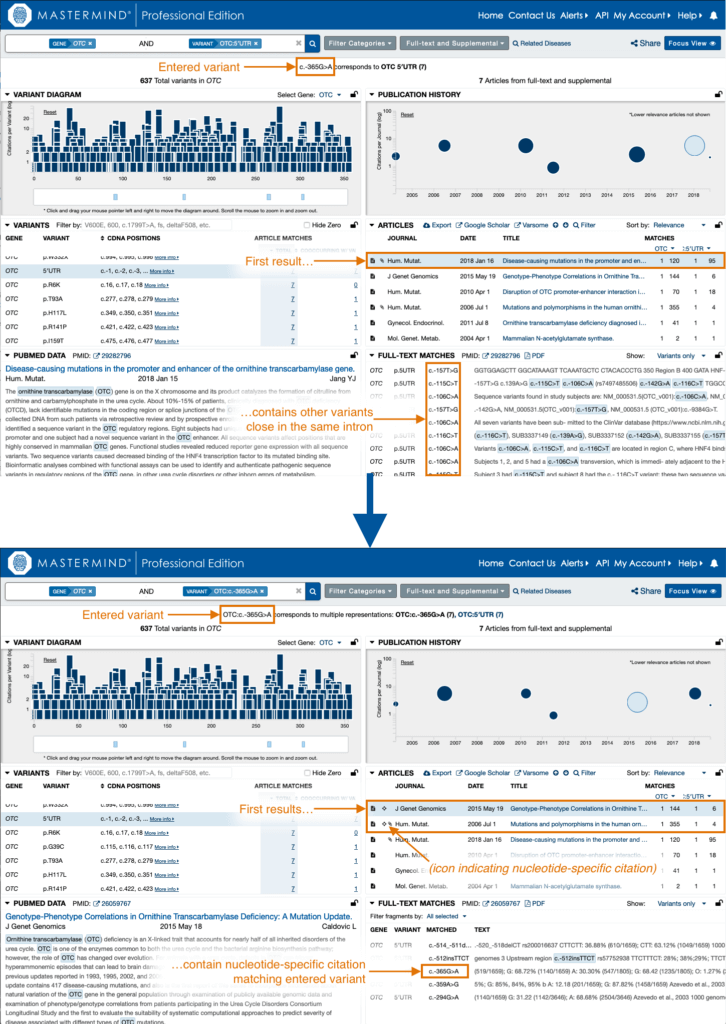
To maximize the practical specificity of results for nucleotide-specific searches without losing the sensitivity of potentially highly-relevant variants, especially in non-coding regions, we now prioritize all articles citing the exact variant searched by nucleotide specificity at the top of the results list.
This way, if nucleotide specificity is desired, you can limit your review to the first articles in the results. Furthermore, if the first article in the list does not match your variant at the nucleotide-specific level, you can be confident that none of the articles in the results will be nucleotide-specific citations.
These results with nucleotide-specific variant citations show a “cross-hair” icon indicating nucleotide precision for the searched variant. Analogously, the “Articles” endpoint in the Advanced API now includes the “matched_dna” property for those same results when searching by a variant with nucleotide-level specificity (using cDNA nomenclature, genomic coordinates, or rsID as input).
See the before and after in the screenshots below.
Splice Region and Intronic Acceptor/Donor Variants
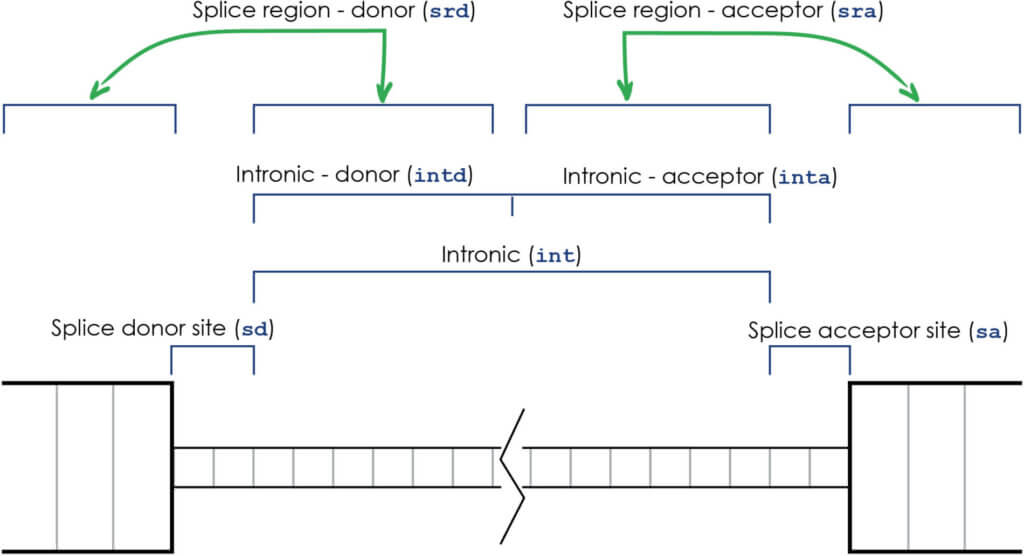
Previously, we’ve divided intronic variants into three groupings:
- sa: the splice acceptor site
- sd: the splice donor site
- int: intronic variants
Intronic variants now have multiple groupings with alternate degrees of specificity, based on biological effect similarities: in addition to the above groupings, we’ve now added:
- srd: splice region donor side variants, which extend from 3 nucleotides into the exon to 8 nucleotides into the intron at the splice donor site
- sra: splice region acceptor side variants, which extend from 3 nucleotides into the exon to 8 nucleotides into the intron at the splice acceptor site
- intd: the donor side of the intron
- inta: the acceptor side of the intron
Introns within Untranslated Regions
We’ve also added additional groupings for variants occurring within intronic regions inside the untranslated regions. So in addition to:
- 5UTR: the 5′UTR region
- 3UTR: the 3′UTR region
Mastermind now includes the old and new intronic variant groupings for intronic variants occurring within 5′UTRs and 3′UTRs as well:
- 5UTRint: intronic variants within the 5′UTR region
- 5UTRintd: intronic donor side variants within the 5′UTR region
- 5UTRinta: intronic acceptor side variants within the 5′UTR region
- 5UTRsd: splice donor site variants within the 5′UTR region
- 5UTRsa: splice acceptor site variants within the 5′UTR region
- 5UTRsrd: splice region donor side variants within the 5′UTR region
- 5UTRsra: splice region acceptor side variants within the 5′UTR region
- 3UTRint: intronic variants within the 3′UTR region
- 3UTRintd: intronic donor side variants within the 3′UTR region
- 3UTRinta: intronic acceptor side variants within the 3′UTR region
- 3UTRsd: splice donor site variants within the 3′UTR region
- 3UTRsa: splice acceptor site variants within the 3′UTR region
- 3UTRsrd: splice region donor side variants within the 3′UTR region
- 3UTRsra: splice region acceptor side variants within the 3′UTR region
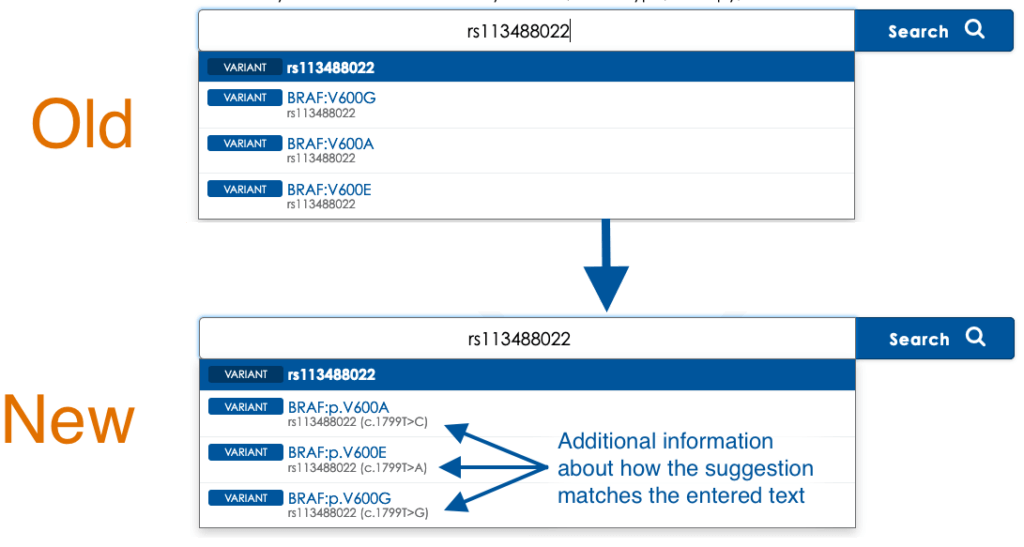
Improved Suggestions for Nucleotide-Specific Searches
When entering an rsID as a search parameter, Mastermind now shows the cDNA nomenclatures which cause the rsID to match each Mastermind grouping.
This information is now provided for genomic coordinate nomenclatures as well.
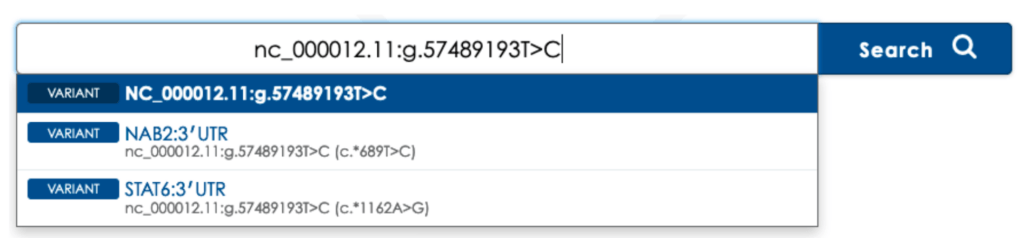
Greater Sensitivity for Overlapping Transcripts
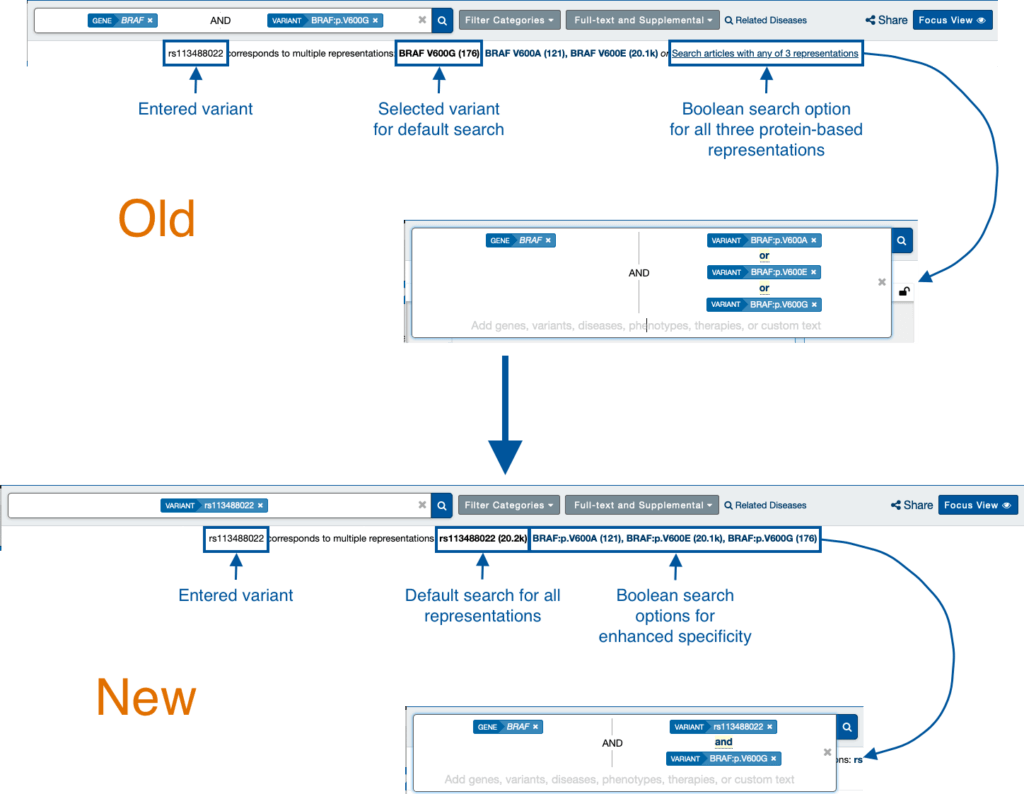
Some variants can correlate to multiple nomenclatures or descriptions resulting from alternate, overlapping transcripts. When starting from a nucleotide-specific nomenclature (cDNA, genomic coordinate, rsID, or IVS), previously you would need to choose the corresponding codon-specific nomenclature (protein-based) to show results, if there were multiple groupings from which to choose.
To maximize sensitivity, in this case, you would need to search each of the corresponding protein-based nomenclatures using Basic Edition, or choose the Boolean search using Professional Edition, to see the entire list of results.
Now, Mastermind will show the fully sensitive results for a nucleotide-specific search by default without requiring Boolean search in Professional Edition.
You still have the option of increasing specificity by filtering down to a particular protein effect, which now retains your nucleotide-specific search as a parameter, and adds the protein-effect as a Boolean filter using Professional Edition.
We hope you find this new approach to improving the precision and prioritization of non-coding variant searches in Mastermind, and welcome your feedback. Visit the Feedback Page.