2018 has been another banner year in the progression of Precision Medicine! Year over year, there has been a dramatic rise in the number of scientific studies and initiatives that resulted in meaningful published findings, and 2018 was another record-breaker. To demonstrate trends in genetics and genomics research seen through the lens of scientific publications, the team mined genomics data aggregated in our Mastermind Genomic Search Engine to reflect on these trends.
The Mastermind Genomic Search Engine has indexed the full text of millions of genomic articles and supplemental data to provide immediate insight into the published research for every disease, gene, and mutation found in the literature. Mastermind is used by hundreds of diagnostic labs around the world to accelerate genomic interpretation and by pharmaceutical companies in drug discovery for its comprehensive genomic landscape of every disease.
New Genomic Insights Indicate an Emergence of Drug Resistance in Cancer and a Focus on Rare Disease Research
Aggregating the granular genomics data allowed us to see the bigger-picture patterns that emerged.
- 2017 findings: Newly-emergent variants were predominantly associated with cancer, and research was predominantly focused on resistance mechanisms or the development of new therapeutics.
- 2018 findings: A continued trend of studies and new discoveries detailing drug resistance mechanisms, with several of the previously identified emerging variants re-appearing on the list of the top 300 newly discovered variants; an increasing number of emergent variants were found in non-cancer diseases. Notable results are summarized in Tables 1 and 2.
Top Emerging Genomic Variants in 2018 – Cancer
Prevalent on this list are variants that drive multiple resistance mechanisms to tyrosine kinase inhibitors: BTK p.C481F, p.C481R, p.C481Y and p.L528W in addition to CCND1 p.E36K that inhibit Ibrutinib; EGFR p. L792H and p.L789I shown to inhibit Osimertinib and Rociletinib, respectively; and resistance variants in ABL1, ALK, ERBB2, NTRK3 and ROS among others (see Table 1).
Highlighting a novel variant of interest discovered in 2018:
- EGFR p.L792H confers resistance to Osimertinib in patients with non-small cell lung cancer (Zhang et al. 2018) and has been found to co-occur with EGFR p.C797S (Yang et al. 2018), as described last year as a newly emerging variant associated with resistance to several tyrosine kinase inhibitors (Avizienyte et al. 2008).
| Gene | Variant | Cancer Type | Resistance | Year First Described | Citation Count | Reference(s) |
|---|---|---|---|---|---|---|
| EGFR | p.L792H | NSCLC | Osimertinib | 2018 | 9 [MM] | Zhang et al. 2018 [PMID: 29857056] |
| AR | p.Q784X | Prostate Cancer | CYP17 inhibitor | 2017 | 5 [MM] | Han et al. 2017 [PMID: 28036278] |
| EGFR | p.L798I | NSCLC | Rociletinib | 2016 | 26 [MM] | Chabon et al. 2016 [PMID: 27283993] |
| NTRK3 | p.G623R | Mammary analogue secretory carcinoma | Entrectinib | 2016 | 21 [MM] | Drilon et al. 2016 [PMID: 26884591] |
| ALK | p.G1202del | NSCLC | Ceritinib, Alectinib, Brigatinib | 2016 | 19 [MM] | Gainor et al. 2016 [PMID: 27432227] Shaw et al. 2017 [PMID: 29074098] |
| CYSLTR2 | L129Q | Uveal melanoma | 2016 | 14 [MM] | Moore et al. 2016 [PMID: 27089179] | |
| ROS | p.S1986Y | NSCLC | Crizotinib | 2016 | 12 [MM] | Facchinetti et al. 2016 [PMID: 27401242] |
| BTK | numerous | CLL | Ibrutinib | 2016 | 8-10 [MM] | Xu et al. 2017 [PMID: 28235842] Sharma et al. 2016 [PMID: 27626698] |
| AR | P.M896V | Prostate Cancer | Enzalutamide | 2016 | 8 [MM] | Prekovic et al. 2018 [PMID: 30306781] |
| ERBB2 | p.K753E | Breast Cancer | Lapatinib | 2016 | 6 [MM] | Zuo et al. 2016 [PMID: 27697991] |
| ABL1 | p.A337V | CML | Asciminib | 2016 | 5 [MM] | Qiang et al. 2017 [PMID: 28819281] |
| CCND1 | p.E36K | MCL | Ibrutinib | 2016 | 5 [MM] | Mohanty et al. 2016 [PMID: 27713153] |
Table 1. Representative emergent variants (newly discovered from 2016 onward) that are associated with cancer and the study of resistance mechanisms or the development of therapeutics. Hyperlinks within citation count show publications for each variant in Mastermind. NSCLC – non-small cell carcinoma. CLL – chronic lymphocytic leukemia. CML – chronic myelogenous leukemia. MCL – mantle cell lymphoma.
Top Emerging Genomic Variants in 2018 – Non-Cancer
Notable Trends:
- A much larger proportion of entries on the list of newly emerging variants were associated with non-malignant diseases (e.g. Thiopurine Intolerance or Obesity)
- A significant percentage of these were associated with rare genetic syndromes, likely reflecting an invigorated interest in studying these less well-characterized diseases using genomic sequencing (e.g. Odgen Syndrome or Activated PI3Kδ Syndrome)
- Some of the notable variants either newly discovered in 2018 or receiving increased attention since their discovery in 2016-2017 are displayed in Table 2 (e.g. LMNA p.R338P causing a form of muscular dystrophy and MEFV p.E244K associated with a form of auto-inflammatory disease)
- Among the emerging variants are variants that cause Activated PI3K-delta Immunodeficiency Syndrome or variants that are strongly associated with influencing body mass index (BMI) such as the variant CREBRF p.R457Q published in Nature Genetics (Minster et al. 2016).
Highlighting some variants of interest:
- HSD17B13 rs72613567 is a loss-of-function variant in HSD17B13 that reduces the risk of non-alcoholic and alcoholic liver disease and cirrhosis as well as reduces the risk of progression from steatosis to steatohepatitis (Abul-Husn et al. 2018).
- ADCY3 c.2433-1G>A disrupts a splice acceptor site causing decreased expression of ADCY3 and is associated with an increased risk of obesity and type II diabetes in the Greenlandic population (Grarup et al. 2018).
| Gene | Variant | Disease/Phenotype | Year First Described | Citation Count | References |
|---|---|---|---|---|---|
| HSD17B13 | rs72613567 | Non-Alcoholic Fatty Liver Disease | 2018 | 8 [MM] | Abul-Husn et al. 2018 [PMID: 29562163] |
| ADCY3 | c.2433–1G>A | Obesity, Type II Diabetes | 2018 | 5 [MM] | Grarup et al. 2018 [PMID: 29311636] |
| PIK3CD | G124D | Activated PI3Kδ Syndrome | 2017 | 10 [MM] | Takeda et al. 2017 [PMID: 28414062] |
| PIK3CD | E81K | Activated PI3Kδ Syndrome | 2017 | 9 [MM] | Takeda et al. 2017 [PMID: 28414062] |
| CYP2R1 | D353D | Vitamin D Deficiency, Multiple Sclerosis | 2017 | 8 [MM] | Manousaki et al. 2017 [PMID: 28757204] |
| PIK3CD | R929C | Hypogammaglobulineamia, Recurrent Infections | 2017 | 8 [MM] | Wentink et al. 2017 [PMID: 28104464] |
| LMNA | R388P | Congenital Muscular Dystrophy, Laminopathy | 2017 | 8 [MM] | Barateau et al. 2017 [PMID: 28125586] |
| MEFV | E244K | Pyrin-Associated Autoinflammation with Neutrophilic Dermatosis | 2017 | 7 [MM] | Moghaddas et al. 2017 [PMID: 28835462] |
| NUDT15 | R139H | Leukopenia, Thiopurine Intolerance | 2016 | 20 [MM] | Moriyama et al. 2016 [PMID: 26878724] |
| CREBRF | R457Q | Obesity | 2016 | 15 [MM] | Minster et al. 2016 [PMID: 27455349] |
| NUDT15 | V18ins | Thiopurine Intolerance | 2016 | 15 [MM] | Moriyama et al. 2016 [PMID: 26878724] |
| GRHL3 | T454M | Nonsyndromic Cleft Palate | 2016 | 13 [MM] | Leslie et al. 2016 [PMID: 27018472] |
| NAA10 | R83C | Ogden Syndrome | 2016 | 12 [MM] | Saunier et al. 2016 [PMID: 27094817] |
| F2 | R596W | Antithrombin Resistance, Venous Thromboembolism | 2016 | 11 [MM] | Bulato et al. 2016 [PMID: 27013614] |
| NAA10 | F128L | Ogden Syndrome | 2016 | 11 [MM] | Saunier et al. 2016 [PMID: 27094817] |
| IL10RA | T179T | Infantile-onset Inflammatory Bowel Disease | 2016 | 11 [MM] | Yanagi et al. 2016 [PMID: 26822028] |
Table 2. A sample of the emergent variants (newly discovered from 2016 onward) in 2018 not associated with cancer. Hyperlinks within citation count show publications for each variant in Mastermind.
A Widening Range of Diseases and Genes of Interest
Taking a step back and looking more broadly at the gene- and disease-level of the newly emergent variants in aggregate, the pattern of emerging research interests as a whole underscored the trends illustrated above: cancer-associated genes for which resistance mechanisms have been identified and genes and diseases associated with rare genetic syndromes dominated the list.
The Top 20 Genes (see Figure 1) and Top 20 Diseases (see Figure 2) most frequently associated with the 300 most emergent variants as published in the 2018 literature were identified in a total of 241 unique genes and 2,228 distinct articles or, on average, 6 to 7 articles per variant. Interestingly…
- The Top 20 Genes associated with the 300 most emergent variants comprised less than 25% of these genes, highlighting the great variety of genes for which newly discovered variants have been described.
- In contrast, the Top 20 Diseases represented about 75% of emerging gene-based research, with approximately 40% of these being malignant and 60% non-malignant.
This result further illustrates the trend towards an increase in rare disease studies focusing on genomic research.
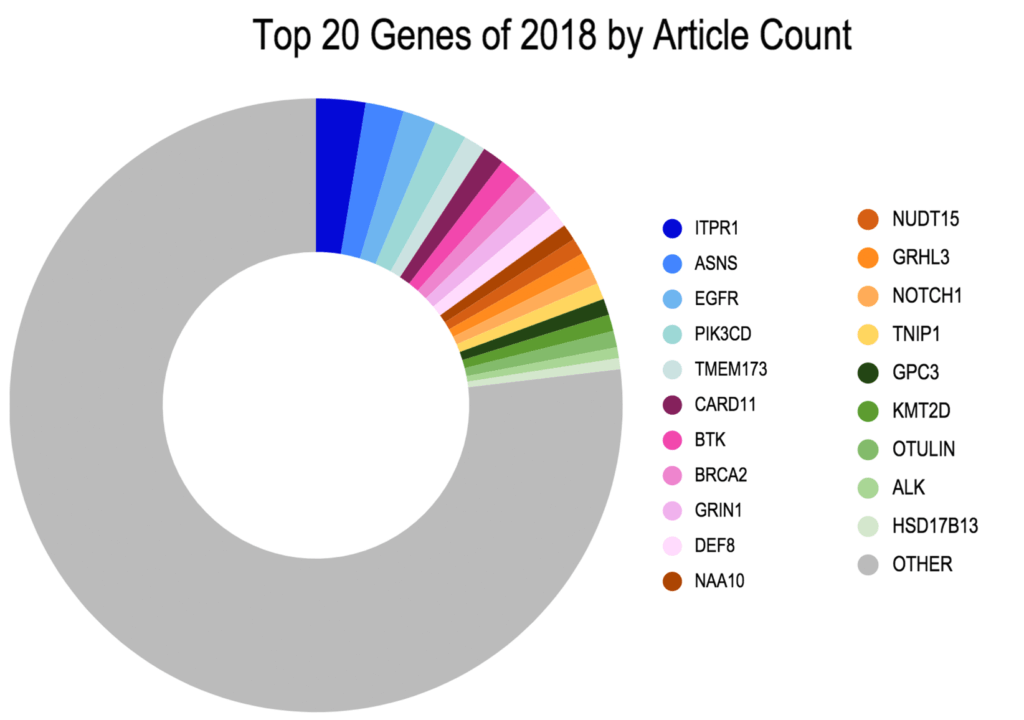
Top 20 Genes of 2018 by Article Count
Figure 1. The Top 20 Genes associated with newly emergent variants as published in the literature between 2016-2018. In total, 241 unique genes were identified with the 300 newly emergent variants.
Interestingly:
- The most widely cited gene associated with newly emergent variants was ITPR1, an inositol1,4,5-triphosphate receptor, associated with spinocerebellar ataxia type 29 (Ando et al. 2018; Casey et al. 2017; Zambonin et al. 2017) or Gillespie Syndrome through both recessive and dominant mechanisms (Gerber et al. 2016).
- The next most widely cited gene on the list was ASNS, the asparagine synthetase gene mutated in the metabolic syndrome ASNS-deficiency first described in a number of case reports in mid to late 2016 and increasingly cited in the literature (Seidahmed et al. 2016; Sun et al. 2016; Yamamoto et al. 2016).
- Rounding out the top five is the stimulator of interferon gene (STING) family member, TMEM173 found to be mutated in STING-associated vasculopathy with onset in infancy (SAVI; Melki et al. 2017) and chilblain lupus (König et al. 2016).
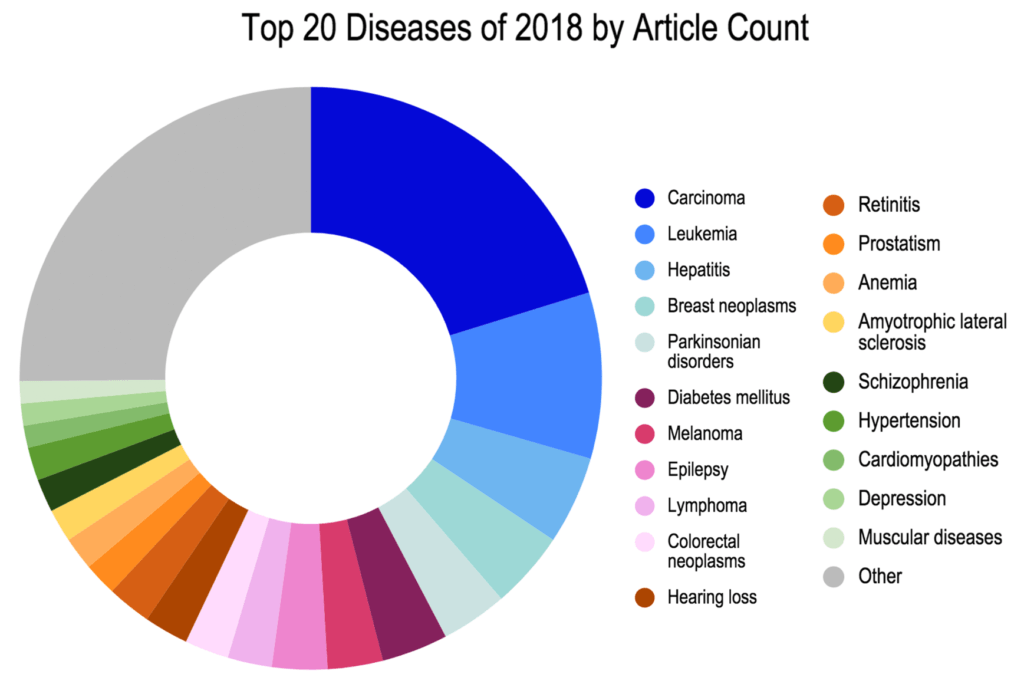
Top 20 Diseases of 2018 by Article Count
Figure 2. The Top 20 Diseases associated with newly emergent variants in literature published from 2016-2018. In total, 48 unique diseases were identified to be primarily associated with the 300 newly emergent variants.
Interestingly:
- A number of newly emerging variants were associated with a variety of different carcinomas and leukemias/lymphomas.
- A synonymous variant in SLC16A11 originally identified in large-scale screening of Mexican populations to identify associations with Type 2 Diabetes was originally published only in supplemental materials (SIGMA Type 2 Diabetes Consortium et al. 2014; Huerta-Chagoya et al. 2015) and is newly emergent as a focus of interest in 2017 articles (Rusu et al.; Miranda-Lora et al.). This variant was found to influence the extent of SLC16A11 transport activity in concert with other associated variants.
- Other highlights include a number of early onset Parkinson’s disease associated variants identified in multiple genes including DNAJC6 p.R927G (Oligatti et al. 2016), PARK7 p.L172Q (Taipa et al. 2016), and SYNJ1 p.R459P (Kirola et al. 2016).
Saturation of 2017-2018 Most-Cited Variants in the Genetic Literature
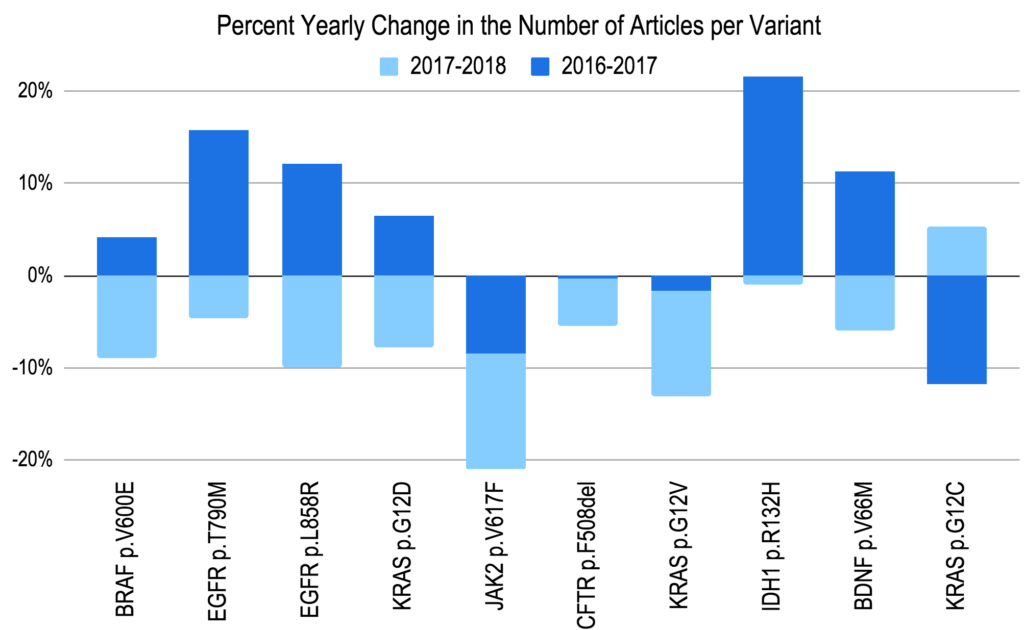
The 2017 study illustrated the nature of the disease-variant landscape by identifying the top 10,000 most widely mentioned clinically significant variants and mapping them over time by publication date. The Top 10 Cited Variants in 2017 were found to be BRAF p.V600E, EGFR p.T790M, JAK2 p.V617F, HFE p.C282Y, EGFR p.L858R, KRAS p.G12D, CFTR p.F508del, KRAS p.G12V, BDNF p.V66M, and SNCA p.A53T (ordered based on most widely mentioned to least mentioned).
Revisiting these data in 2018, all of the previously top cited variants experienced a decrease in the number of publications in 2018, with some variants having already experienced a decrease from 2016 to 2017 (see Figure 3 for more details). This surprising trend suggests that these widely published variants may be reaching a saturation point in the genetic literature. In other words, the variants are well-studied enough that publication rates have begun to decline while other newly discovered variants for a more diverse collection of genes and diseases are getting more attention.
Top 10 Cited Variants in 2018 as Compared to 2017 [Percent Yearly Change]
Figure 3. The top 10 most widely cited variants of 2017, versus the percent change in the number of citing articles over two years: 2016-2017 (dark) and 2017-2018 (light).
Genomic Research Takes on a New Direction
By utilizing the Mastermind database, an increasingly diverse array of newly emergent variants has been discovered and described in the scientific literature.
This data showcases a shift of focus away from previously well-characterized variants that have been known for many years, and from a singular focus on cancer towards the inclusion of an increasingly broad array of variants in many more types of genes and a greater number of constitutional diseases, including rare genetic disorders.
As such, the newly emergent cancer-associated variants tend to be associated with resistance mechanisms. This insight highlights the likelihood that they are newly appearing in nature as opposed to being newly discovered (as would be expected of the variants associated with constitutional diseases, for which the absence of treatment pressures does not lead to disease evolution in the form of new genetic variation). Discovering and characterizing these drug resistance mechanisms in cancer will likely remain a prominent component of genomic research for the foreseeable future.
The analysis, as performed within this review allowed us to uncover interesting epidemiological patterns as they emerge. It will be interesting to see how these trends expand and evolve in future years.
For clinical practice, the recognition of these new resistance mechanisms in cancer and newly discovered disease-causing variants in rare disease will surely inform patient care, and highlights the need for practitioners to stay abreast of the literature much more faithfully than in years past.
Overall, the data presented here suggests that the diversifying landscape of genetic research foreshadows a substantial increase in the development of personalized medicine for rare diseases and drug resistance in the coming years.
Get a printable copy of this report
Read our next report: Fusion Genes of Clinical Significance in 2019
The content as presented in this article is a current variant, gene, and disease trends analysis as of January 2019, and provided by Genomenon team members Nathan Adams, Lauren Chunn, Mark Kiel, and Diane Nefcy. This study was also published in EnlightenBio in collaboration with Brigitte Ganter.
Mastermind, Genomenon’s Genomic Search Engine, provides immediate insight into the published genomic research for every disease, gene, and genetic variant found in the literature. Used by hundreds of diagnostic labs around the world, Mastermind accelerates genomic interpretation by providing unique insight into genomic relationships found in the full text of millions of scientific articles. Pharmaceutical researchers license the Mastermind database for a comprehensive genomic landscape associated with any given disease – to identify and prioritize genomic biomarkers for drug discovery and clinical trial targets. Mastermind Alerts provides automatic notifications of new articles citing requested genes, variants, and diseases on a weekly basis.
References
Abul-husn NS, Cheng X, Li AH, et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med. 2018;378(12):1096-1106. [PMID: 29562163]
Ando H, Hirose M, Mikoshiba K. Aberrant IP receptor activities revealed by comprehensive analysis of pathological mutations causing spinocerebellar ataxia 29. Proc Natl Acad Sci USA. 2018;115(48):12259-12264. [PMID: 30429331]
Avizienyte E, Ward RA, Garner AP. Comparison of the EGFR resistance mutation profiles generated by EGFR-targeted tyrosine kinase inhibitors and the impact of drug combinations. Biochem J. 2008;415(2):197-206. [PMID: 18588508]
Barateau A, Vadrot N, Vicart P, et al. A Novel Lamin A Mutant Responsible for Congenital Muscular Dystrophy Causes Distinct Abnormalities of the Cell Nucleus. PLoS ONE. 2017;12(1):e0169189. [PMID: 28125586]
Bulato C, Radu CM, Campello E, et al. New Prothrombin Mutation (Arg596Trp, Prothrombin Padua 2) Associated With Venous Thromboembolism. Arterioscler Thromb Vasc Biol. 2016;36(5):1022-9. [PMID: 27013614]
Casey JP, Hirouchi T, Hisatsune C, et al. A novel gain-of-function mutation in the ITPR1 suppressor domain causes spinocerebellar ataxia with altered Ca signal patterns. J Neurol. 2017;264(7):1444-1453. [PMID: 28620721]
Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. [PMID: 27283993]
Drilon A, Li G, Dogan S, et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol. 2016;27(5):920-6. [PMID: 26884591]
Facchinetti F, Loriot Y, Kuo MS, et al. Crizotinib-Resistant ROS1 Mutations Reveal a Predictive Kinase Inhibitor Sensitivity Model for ROS1- and ALK-Rearranged Lung Cancers. Clin Cancer Res. 2016;22(24):5983-5991. [PMID: 27401242]
Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6(10):1118-1133. [PMID: 27432227]
Gerber S, Alzayady KJ, Burglen L, et al. Recessive and Dominant De Novo ITPR1 Mutations Cause Gillespie Syndrome. Am J Hum Genet. 2016;98(5):971-980. [PMID: 27108797]
Grarup N, Moltke I, Andersen MK, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018;50(2):172-174. [PMID: 29311636]
Han D, Gao S, Valencia K, et al. A novel nonsense mutation in androgen receptor confers resistance to CYP17 inhibitor treatment in prostate cancer. Oncotarget. 2017;8(4):6796-6808. [PMID: 28036278]
Huerta-chagoya A, Vázquez-cárdenas P, Moreno-macías H, et al. Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women. PLoS ONE. 2015;10(5):e0126408. [PMID: 25973943]
Kirola L, Behari M, Shishir C, Thelma BK. Identification of a novel homozygous mutation Arg459Pro in SYNJ1 gene of an Indian family with autosomal recessive juvenile Parkinsonism. Parkinsonism Relat Disord. 2016;31:124-128. [PMID: 27496670]
König N, Fiehn C, Wolf C, et al. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann Rheum Dis. 2017;76(2):468-472. [PMID: 27566796]
Leslie EJ, Liu H, Carlson JC, et al. A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. Am J Hum Genet. 2016;98(4):744-54. [PMID: 27018472]
Manousaki D, Dudding T, Haworth S, et al. Low-Frequency Synonymous Coding Variation in CYP2R1 Has Large Effects on Vitamin D Levels and Risk of Multiple Sclerosis. Am J Hum Genet. 2017;101(2):227-238. [PMID: 28757204]
Melki I, Rose Y, Uggenti C, et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J Allergy Clin Immunol. 2017;140(2):543-552.e5. [PMID: 28087229]
Minster RL, Hawley NL, Su CT, et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat Genet. 2016;48(9):1049-1054. [PMID: 27455349]
Miranda-lora AL, Cruz M, Molina-díaz M, Gutiérrez J, Flores-huerta S, Klünder-klünder M. Associations of common variants in the SLC16A11, TCF7L2, and ABCA1 genes with pediatric-onset type 2 diabetes and related glycemic traits in families: A case-control and case-parent trio study. Pediatr Diabetes. 2017;18(8):824-831. [PMID: 28101933]
Moghaddas F, Llamas R, De nardo D, et al. A novel Pyrin-Associated Autoinflammation with Neutrophilic Dermatosis mutation further defines 14-3-3 binding of pyrin and distinction to Familial Mediterranean Fever. Ann Rheum Dis. 2017;76(12):2085-2094. [PMID: 28835462]
Mohanty A, Sandoval N, Das M, et al. CCND1 mutations increase protein stability and promote ibrutinib resistance in mantle cell lymphoma. Oncotarget. 2016;7(45):73558-73572. [PMID: 27713153]
Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48(6):675-80. [PMID: 27089179]
Moriyama T, Nishii R, Perez-andreu V, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48(4):367-73. [PMID: 26878724]
Olgiati S, Quadri M, Fang M, et al. DNAJC6 Mutations Associated With Early-Onset Parkinson’s Disease. Ann Neurol. 2016;79(2):244-56. [PMID: 26528954]
Prekovic S, Van den broeck T, Linder S, et al. Molecular underpinnings of enzalutamide resistance. Endocr Relat Cancer. 2018;25(11):R545–R557. [PMID: 30306781]
Qiang W, Antelope O, Zabriskie MS, et al. Mechanisms of resistance to the BCR-ABL1 allosteric inhibitor asciminib. Leukemia. 2017;31(12):2844-2847. [PMID: 28819281]
Rusu V, Hoch E, Mercader JM, et al. Type 2 Diabetes Variants Disrupt Function of SLC16A11 through Two Distinct Mechanisms. Cell. 2017;170(1):199-212.e20. [PMID: 28666119]
Saunier C, Støve SI, Popp B, et al. Expanding the Phenotype Associated with NAA10-Related N-Terminal Acetylation Deficiency. Hum Mutat. 2016;37(8):755-64. [PMID: 27094817]
Seidahmed MZ, Salih MA, Abdulbasit OB, et al. Hyperekplexia, microcephaly and simplified gyral pattern caused by novel ASNS mutations, case report. BMC Neurol. 2016;16:105. [PMID: 27422383]
Sharma S, Galanina N, Guo A, et al. Identification of a structurally novel BTK mutation that drives ibrutinib resistance in CLL. Oncotarget. 2016;7(42):68833-68841. [PMID: 27626698]
Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017;18(12):1590-1599. [PMID: 29074098]
Sun J, Mcgillivray AJ, Pinner J, et al. Diaphragmatic Eventration in Sisters with Asparagine Synthetase Deficiency: A Novel Homozygous ASNS Mutation and Expanded Phenotype. JIMD Rep. 2017;34:1-9. [PMID: 27469131]
Taipa R, Pereira C, Reis I, et al. DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain. 2016;139(Pt 6):1680-7. [PMID: 27085187]
Takeda AJ, Zhang Y, Dornan GL, et al. Novel PIK3CD mutations affecting N-terminal residues of p110δ cause activated PI3Kδ syndrome (APDS) in humans. J Allergy Clin Immunol. 2017;140(4):1152-1156.e10. [PMID: 28414062]
Wentink M, Dalm V, Lankester AC, et al. Genetic defects in PI3Kδ affect B-cell differentiation and maturation leading to hypogammaglobulineamia and recurrent infections. Clin Immunol. 2017;176:77-86. [PMID: 28104464]
Williams AL, Jacobs SB, Moreno-macías H, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506(7486):97-101. [PMID: 24390345]
Xu L, Tsakmaklis N, Yang G, et al. Acquired mutations associated with ibrutinib resistance in Waldenström macroglobulinemia. Blood. 2017;129(18):2519-2525. [PMID: 28235842]
Yamamoto T, Endo W, Ohnishi H, et al. The first report of Japanese patients with asparagine synthetase deficiency. Brain Dev. 2017;39(3):236-242. [PMID: 27743885]
Yanagi T, Mizuochi T, Takaki Y, et al. Novel exonic mutation inducing aberrant splicing in the IL10RA gene and resulting in infantile-onset inflammatory bowel disease: a case report. BMC Gastroenterol. 2016;16:10. [PMID: 26822028]
Yang Z, Yang N, Ou Q, et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018;24(13):3097-3107. [PMID: 29506987]
Zambonin JL, Bellomo A, Ben-pazi H, et al. Spinocerebellar ataxia type 29 due to mutations in ITPR1: a case series and review of this emerging congenital ataxia. Orphanet J Rare Dis. 2017;12(1):121. [PMID: 28659154]
Zhang Q, Zhang XC, Yang JJ, et al. EGFR L792H and G796R: Two Novel Mutations Mediating Resistance to the Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib. J Thorac Oncol. 2018;13(9):1415-1421. [PMID: 29857056]
Zuo WJ, Jiang YZ, Wang YJ, et al. Dual Characteristics of Novel HER2 Kinase Domain Mutations in Response to HER2-Targeted Therapies in Human Breast Cancer. Clin Cancer Res. 2016;22(19):4859-4869. [PMID: 27697991]