Original version of this post was previously published on the Enlightenbio Blog on January 1, 2018 by Brigitte Ganter.
Dr. Mark Kiel (CSO and co-Founder at Genomenon) and Lauren Chunn (Data Processing Analyst at Genomenon) were able to mine the Mastermind database of genomic variants and analyze the trending citations of widely covered variants in the medical literature – including one of the most notable variants: EGFR p.C797S.
Through this process, they paint the picture of the changing landscape of genomic research and medicine, from variants that have remained a common feature for decades to newly emerging variants over the last few years.
Mastermind Genomic Database
Mastermind, a comprehensive database of genomic disease-to-gene-to-variant associations, supports its users to search through millions of full-text articles from the primary medical literature to identify variants of interest, prioritize them, and retrieve relevant articles for disease-gene-variant combinations. The richness of Mastermind’s content and the associated functionalities can retrieve interesting citation data for analysis.
Mastermind is being used by hundreds of laboratories and over 500 users across 20 countries—either as the Free Edition of Mastermind or via subscription to Mastermind Professional Edition—for enhancing and automating high-throughput workflows.
2017 Top Ten Variants
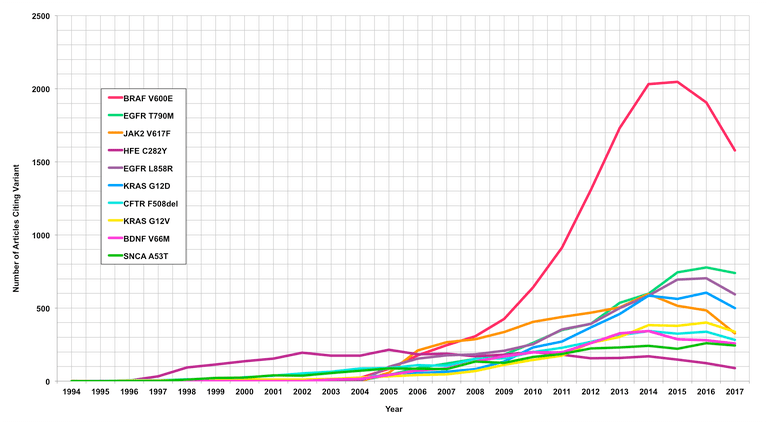
To illustrate the nature of the disease-to-gene-to-variant landscape, the top 10,000 most widely mentioned, clinically significant variants were identified and mapped over time by publication date.
With the exception of BRAF:V600E, the top ten most widely cited variants, including:
EGFR:T790M, JAK2:V617F, HFE:C282Y, EGFR:L858R, KRAS:G12D, CFTR:F508del, KRAS:G12V, BDNF:V66M, and SNCA:A53T
Exhibit a steady upward curve in the number of citations over time as shown in Figure 1.
Not surprisingly, the cancer associated with BRAF:V600E mutation remains the most highly cited variant from 2008 onward, with a substantial increase in the number of citations between 2008 and 2014. Furthermore, of the top ten highly cited variants in total, six are associated with cancer as might be expected, whereas four are associated with constitutional diseases, including Parkinson’s disease, hemochromatosis, and cystic fibrosis.
Figure 1: Shown are the top ten most widely cited variants graphed as the number of citations per year. Analysis time frame: 1994-2017.
Newly emerging, highly cited variants are associated with cancer, with a focus on resistance mechanisms and the development of new therapeutics
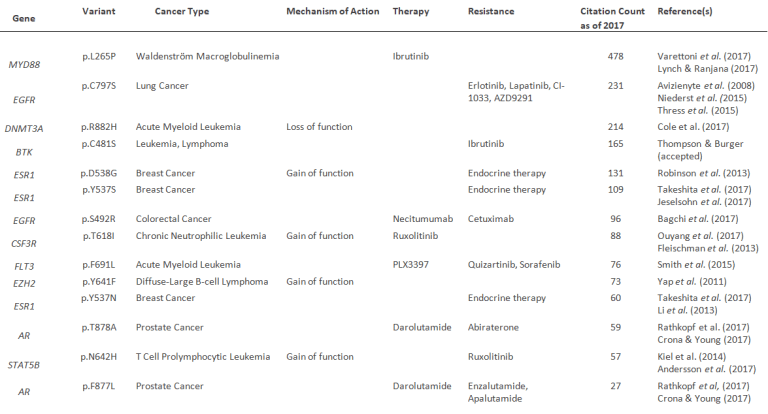
While the top ten identified variants illustrate the direction and focus of genomic research over several past decades, a second picture emerges, demonstrating that these variants are predominantly associated with cancer and, more specifically, with a focus on resistance mechanisms and the development of new therapeutic interventions as summarized in the table below.
Table 1: Cancer-associated variants that have greatly increased in the number of citations in the past few years and are relevant to resistance mechanisms and/or therapeutic developments.
EGFR p.C797S mutation details
One of the most notable variants is EGFR p.C797S, a mutation acquired only after treatment with EGFR tyrosine kinase inhibitors (TKIs), which confers resistance to these drugs in the treatment of lung cancer (Avizienyte et al., 2008).
EGFR p.C797S mutation details
- This specific variant was first reported in November of 2007 by Yu et al.
- The first documentation of its role in resistance occurred in October of 2008 by Avizienyte et al. (AstraZeneca).
- Avizienyte and team found that the presence of the C797S mutation on the same EGFR allele as another mutation, p.T790M (one of the most widely cited variants overall), resulted in complete resistance to erlotinib, lapatinib, and the investigational drug CI-1033 (Avizienyte et al., 2008).
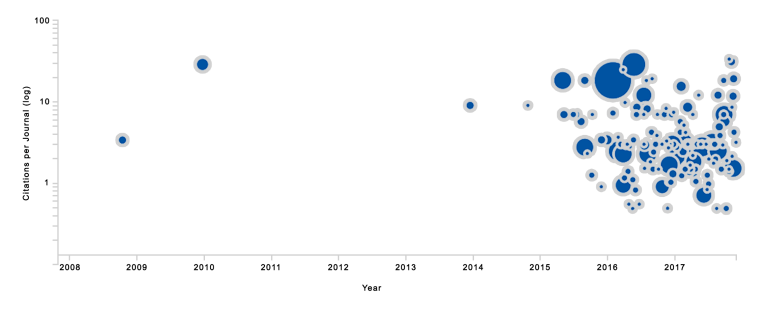
- The variant was mentioned in the scientific literature only five times until 2015.
- However, since 2015, the variant has been discussed in 218 articles (see also Figure 2).
- The 2015 article by Niederst et al. (2008), Massachusetts General Hospital Center, described that when the p.C797S and p.T790M mutations occur in trans, or on different alleles, the cells remain resistant to third generation EGFR TKIs, but are sensitive to a combination of first and third generation EGFR Coinciding with the findings of that previous article is the observation that when those mutations occur in cis, or on the same allele, the cells remain fully resistant to EGFR TKIs used alone or in combination (Niederst et al., 2008).
- Additional variants with precision therapeutic implications are depicted in Table 1, along with their disease associations and therapeutic affinities.
Figure 2: Mutation EGFR p.C797S graphed as the number of citations per year (log scale). Each bubble represents a single article with the bubble size reflecting its relevance to genomic medicine (as determined by a quantification algorithm).
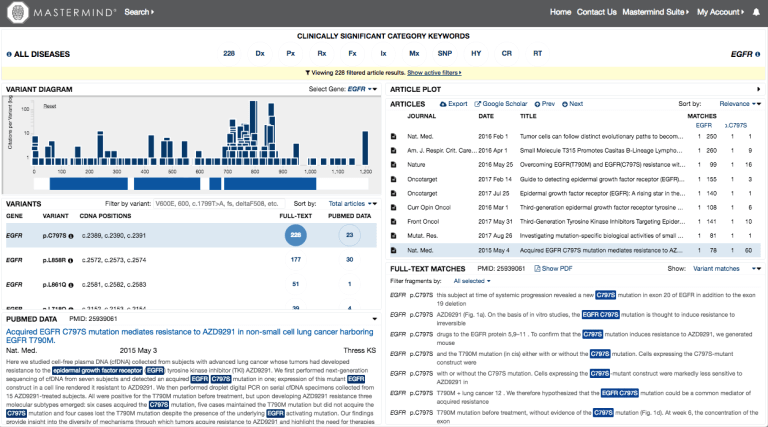
Figure 3: Mastermind software screenshot highlighting the references identifying the EGFR p.C797S variant.
An upward trend toward constitutional diseases in parallel to ongoing cancer research
The EGFR p.C797S mutation is an excellent example of the current direction of genomic research focusing on cancer, precision therapies, and drug resistance mechanisms. However, many of the variants experiencing a similar burst in citations within the last few years are found to be involved in constitutional diseases (generally defined as pathological lesions whose etiology depends to a significant degree upon the action of genetic factors), particularly Alzheimer disease/dementia and Parkinson’s disease.
Genes and variants identified with greatly increased presence in the genomic literature include:
- With an increasing focus on neurodegenerative disease, neuropsychiatric, and other disorders:
- PARK2, VPS35, and SNCA: variants in three genes implicated in the pathogenesis of Parkinson’s disease
- MAPT and TREM2: variants in genes implicated in the risk for and prognosis of Alzheimer disease
- NUDT15 R139C: a variant associated with thiopurine-induced hair loss and leukopenia in the treatment of inflammatory bowel disease (Kakuta et al.,2016).
- SLC10A1 S267F: a variant associated with a decreased risk of cirrhosis and hepatocellular carcinoma in patients with chronic hepatitis B infection (Hu et al., 2016)
- Other notable variants, some of which are associated with cancer, include:
- RAD51 G151D: a variant associated with a novel hyper-recombination phenotype and resistance to select DNA damaging agents (Marsden et al., 2016)
- MITF E318K: a germ-line variant associated with an increased risk of developing melanoma (Potrony et al., 2016)
- SF3B1 K700E: a variant associated with impaired erythropoiesis in myelodysplastic syndrome (Obeng et al., 2016)
Genomenon, through the use of its Mastermind database, was able to demonstrate how the genomic literature has been changing over the recent years, from the consistency of research involving widely known variants such as BRAF p.V600E to the substantial bursts of research describing newly discovered variants. Furthermore, it’s clear that the current field of genomic research is skewed toward cancer research with a recent focus on the development of targeted therapeutics. However, while cancer research seems to dominate, the recent trend towards increased publication of variants affecting constitutional diseases such as Alzheimer and Parkinson’s diseases may foreshadow the emergence of personalized therapies for treatment of such diseases.