By allowing users to find, connect, explore, and understand the links between genomic concepts of interest, the Mastermind Genomic Search Engine has quickly become the essential variant interpretation literature search companion. In this blog, we explore Mastermind’s utility for finding data on gene fusions.
Compelling data in support of this claim was recently published by our team in a peer-reviewed paper in Frontiers in Genetics entitled Mastermind: A Comprehensive Genomic Association Search Engine for Empirical Evidence Curation and Genetic Variant Interpretation. This paper illustrates how our AI-driven solution has automatically identified and annotated over 6.8 million unique genetic variants (the number is now more than 9.6 million) across the published genetic evidence – a more than 22-fold increase compared to the largest manually curated alternative.
Though the yield of genetic variants is both interesting and highly valuable for resolving VUSs, Mastermind can also deliver profound insight around other genomic phenomena – particularly, fusion genes.
A Review of Gene Fusions
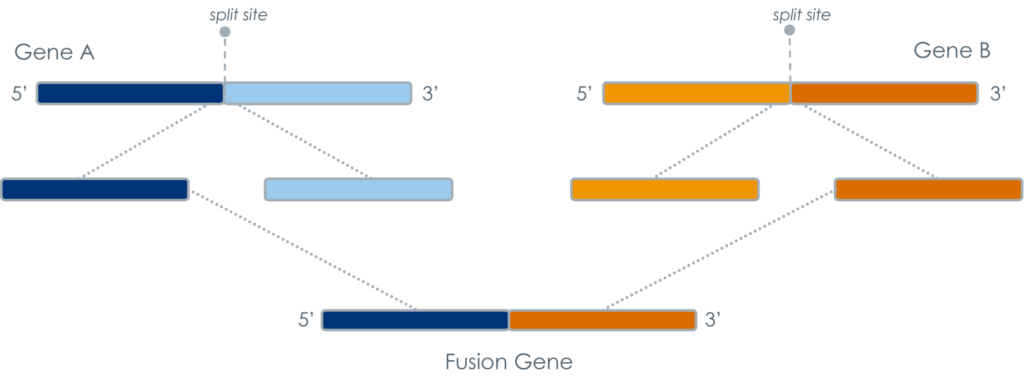
In early stages of gene formation, chromosomes from two different genes may rearrange or translocate, forming a fusion gene. Moving forward, pieces of two genes are essentially processed as a single unit, which may lead to the generation of a dysfunctional protein. Much like vehicle assembly, if two different cars have a few components switched with each other early in the process, the likelihood of functional issues increases.
Though researchers may initiate these “molecular mistakes” in vitro for research purposes, fusion events of particular interest are ones that can occur naturally in the body. In fact, abnormal fusion genes and their associated proteins often play a key role in a variety of cancers.
Fusion Genes and Cancer
Since the discovery of the first fusion gene in 1960, researchers have continued to explore and discover fusion events involved in cancer development. In the past decades, thousands of novel fusions have been identified across a variety of cancer types. This is largely due to the ease of producing this data using next generation sequencing (NGS) technology, and their broad use in clinical diagnostic labs.
Identifying and documenting each newly discovered fusion is crucial in both patient diagnosis and the development of precision medicine. This diagnostic method allows for the proper application of existing therapies and the development of new therapies.
The first known fusion gene, BCR-ABL1, was discovered as an abnormally small chromosome during the analysis of chronic myelogenous leukemia (CML) cells. Over a decade later, it was later characterized as an abnormal translocation of chromosomes 9 and 22. Then, in the 1980s, it was understood to cause the fusion of two gene products – BCR and ABL1. Now, this fusion gene is known to be present in more than 95% of CML patients and 5-25% of acute lymphoblastic leukemia patients [1-3].
Over the years, fusion genes have become an increasingly important therapeutic target. In the case of the BCR–ABL1 fusion gene, isolating the mechanism of dysfunction (overactive enzyme activity) allowed for the development of a highly effective precision targeted therapy that led to dramatic reductions in disease burden with minimal off-target effects.
Since then, other successful drug programs targeting fusion genes have been promulgated, as they represent a significant proportion of cancer patients. A 2017 study reported that 15% of patients with metastatic cancer harbored genomic rearrangements, many of which spawned gene fusions [4]. While 35% of these fusions involved kinase genes and may respond to currently existing kinase inhibitors, 19% of them involved novel partner genes. This discovery left open the possibility for new strategies of drug development [4]. In a large-scale 2018 study, a total of 25,000 fusions were discovered across 9,600 tumor samples encompassing 33 different cancer types. Fusion events drove pathogenesis of 16.5% of these cancers, and were the sole driver in 1.8% of them. Additionally, 6% of the samples contained fusions that could potentially be targeted by currently existing therapies [5].
Mastermind as a Solution for Fusion Documentation
Many fusions discovered in tumor samples involve novel partner genes, and these fusions can often be targeted by existing or emerging therapies. Currently, the Catalogue of Somatic Mutations in Cancer (COSMIC) and The Cancer Genome Atlas (TCGA) serve as the main source for documented fusions [6]. COSMIC contains 297 unique fusion pairs derived from ~1.4 million tumor samples, while TCGA contains clinical and sequencing data from over 20,000 samples [6].
We tested whether Mastermind could serve as a more comprehensive source for documented fusions, and developed a process to automatically retrieve fusion genes from our database of full-text genomic articles. To focus our study on fusion events of clinical significance, we restricted our analysis to the 507 genes comprising the Illumina TruSight Fusion Gene Panel.
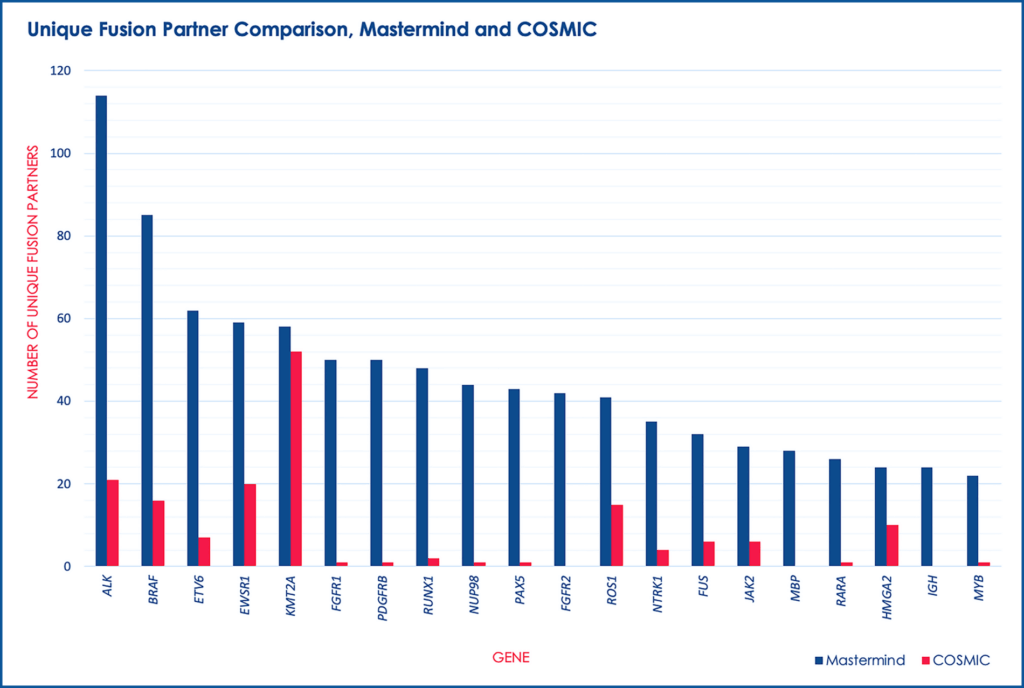
For these 507 genes, we discovered almost 2,000 unique fusion pairs cited in the scientific literature, all of which were manually validated. This represents a 538% increase in yield over the 297 unique fusions in COSMIC. Additionally, we identified numerous published clinically significant fusions that are not found in neither COSMIC nor TCGA.
Unique fusion partner comparison, Mastermind and COSMIC.
Our results indicate COSMIC is an insufficient catalogue of gene fusion events for interpretation of molecular results from clinical samples. Specifically, COSMIC lacks the literature support needed to ensure that the database is fully inclusive of all documented fusions, as evidenced by fusions Mastermind identified that were present in neither database. Additionally, the fusions in COSMIC are aggregated from large sequencing studies, which prevents the more detailed curation that is possible through analysis of the literature.
Mastermind and the Future of Gene Fusions
Using Mastermind, we were able to discover several-fold more fusions than were documented in the COSMIC database. This comprehensive understanding of the complete gene fusion landscape as published in the medical literature is vital to ensuring accurate diagnosis and treatment of cancer patients whose disease is caused by fusion events. As publication of gene fusions and their function in the development of cancer continues to increase, identifying and documenting each newly discovered fusion will only become more crucial in both patient diagnosis and the development of precision medicine. Ultimately, a deeper understanding of fusion genes not only enriches existing therapies, but also stimulates the development of new ones.
With Genomenon’s AI-driven genomic engine, a comprehensive view of the fusion landscape in cancer can be developed by indexing the entire body of medical evidence. The resulting gene fusion database can provide insight into patient diagnosis and treatment decisions, and provide a platform for both drug discovery and repurposing efforts.
Mastermind, with its ability to systematically organize and analyze the medical/genetic literature, can serve as a more comprehensive source for the documentation of fusion events in both constitutional diseases and somatic cancer.
These actionable insights around fusion genes represent a small piece of Mastermind’s capabilities. To see what it can reveal for your complex genetic data, sign up for your free Mastermind account to begin exploring. There’s so much more to discover.
References
-
-
- Parker, Brittany C., and Zhang, Wei. “Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment.” Chinese journal of cancer vol. 32,11 (2013): 594-603.
- Orna, Dreazen et al. “Multiple molecular abnormalities in Ph1 chromosome positive acute lymphoblastic leukaemia.” British Journal of Haematology vol. 67,11 (1987): 319-324.
- Nowell, P. C. and Hungerford, D. A. “A minute chromosome in human chronic granulocytic leukemia.” Science 132, (1960): 1497.
- Zehir, Ahmet et al. “Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients.” Nature medicine vol. 23,6 (2017): 703-713.
- Gao, Qingsong et al. “Driver Fusions and Their Implications in the Development and Treatment of Human Cancers.” Cell reports vol. 23,1 (2018): 227-238.
- Tate, John G. et al. “COSMIC: the Catalogue Of Somatic Mutations In Cancer.” Nucleic acids research vol. 47,D1 (2018): D941-D947.
-