Mastermind uses shorthand identifiers to represent different types of variants, which are displayed and searchable within the Variants table. The shorthands used by Mastermind are as follows:
- Insertions: “ins” — e.g. V600ins
- Deletions: “del” — e.g. V600del
- Indels: “delins” — e.g. V600delins
- Nonsense: “X” — e.g. V600X
- Frameshift: “fs” — e.g. V600fs
- Untranslated regions: “UTR” — e.g. 5’UTR or 3’UTR. Some genes contain introns within the untranslated regions; therefore a variant might belong to both the “UTR” and “int” groups simultaneously – e.g. 5’UTRint.
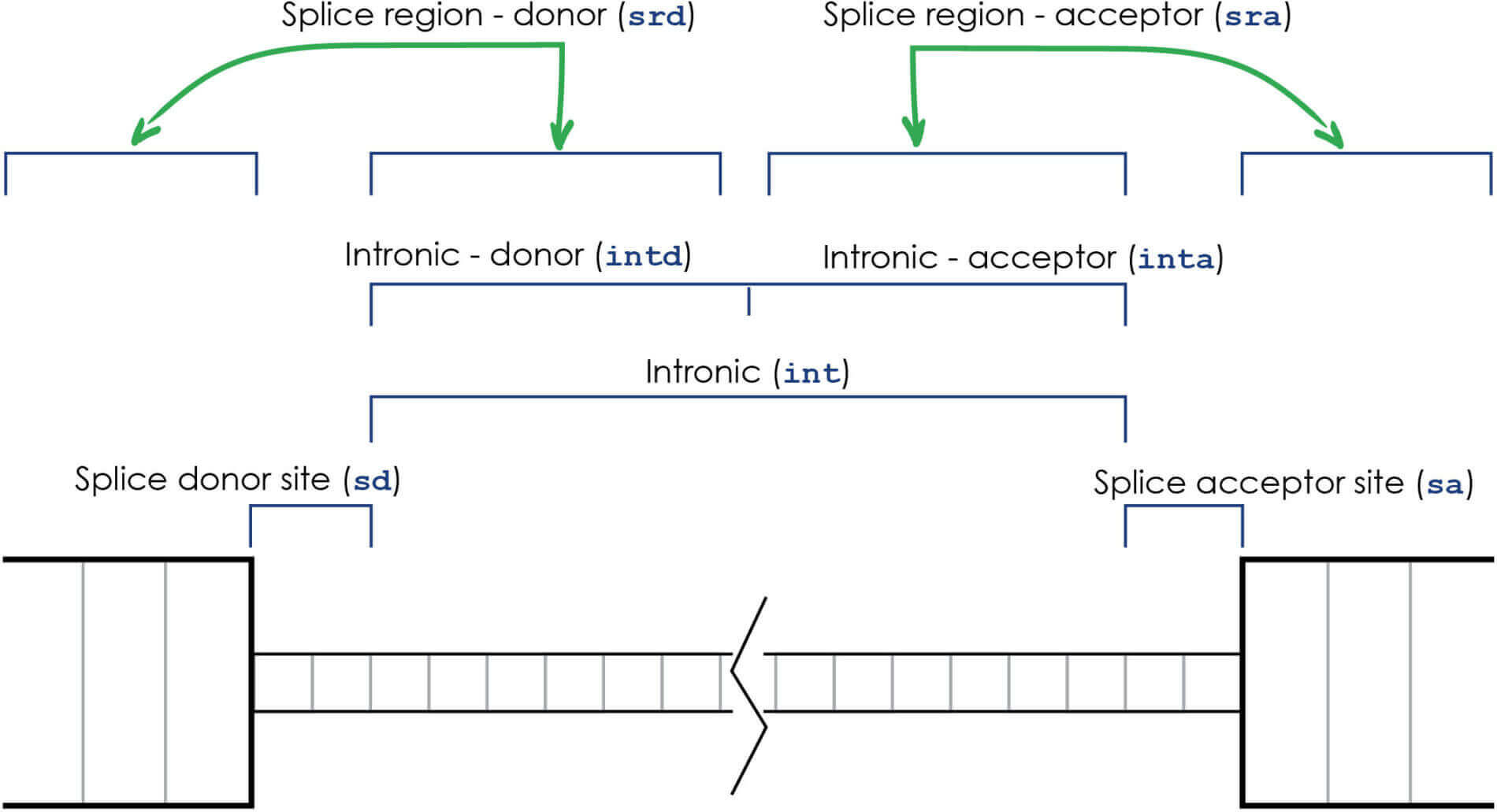
- Splice donor: “sd” — e.g. V168sd; these are variants affecting the 2-base region at the 5′ side of the intron. In the protein space, these are mapped to the nearest amino acid in the nearest coding neighbor at the 5′ side of the intron.
- Splice acceptor: “sa” — e.g. N581sa; these are variants affecting the 2-base region at the 3′ side of the intron. In the protein space, these are mapped to the nearest amino acid in the nearest coding neighbor at the 3′ side of the intron.
- Intronic: “int” — e.g. E46int; these are variants affecting any of the bases within the intron between the splice acceptor and splice donor sites. In the protein space, these are mapped to the nearest amino acid in the nearest coding neighbor.
- Intronic donor and acceptor sides: “intd” and “inta” — e.g. N581intd or N581inta; these are variants that occur in either the donor half or the acceptor half of the Intronic “int” variant region between the splice donor and splice acceptor sites. These are more specific sub-divisions of the “int” category, and so variants in either the “intd” or “inta” categories will appear in the “int” category as well.
- Splice regions: “srd” and “sra” — e.g. N581srd or N581sra; these are variants surrounding the splice sites, from 1 to 3 bases into the exon and from 3 to 8 bases into the intron. The intronic part of the splice regions overlap the intronic “int” classification as well, so splice region variants within the intron will also appear in the “int” and either the “intd” or “inta” categories as well.
- Upstream genetic variant: “ugv” — these are variants affecting the region of 5,000 bases upstream of the 5′ side of the gene.
- Downstream genetic variant: “dgv” — these are variants affecting the region of 5,000 bases downstream of the 3′ side of the gene.
- Extensions: “ext” — e.g. M1ext; these are variants extending the reference amino acid sequence at the N- or C-terminal end with one or more amino acids.
The graphic below shows the different regions for these groups of variants: