On 06/26/25, Genomenon hosted a dynamic and highly informative webinar, "Real World Evidence for Precision Therapeutics," showcasing how expertly curated genomic and biological data can transform drug development and rare disease diagnosis. Moderated by Genomenon's Chief Science Officer and Founder, Dr. Mark Kiel, the session featured compelling presentations from Jonathan Eads (VP of Technology at Genomenon) and Dr. Sarah Chang (Medical Strategy Lead at UCB). The conversation provided a clear, actionable understanding of how real-world data, powered by genomic intelligence, is reshaping the landscape for both rare disease and oncology applications.
The Big Picture: What We Learned from the Experts
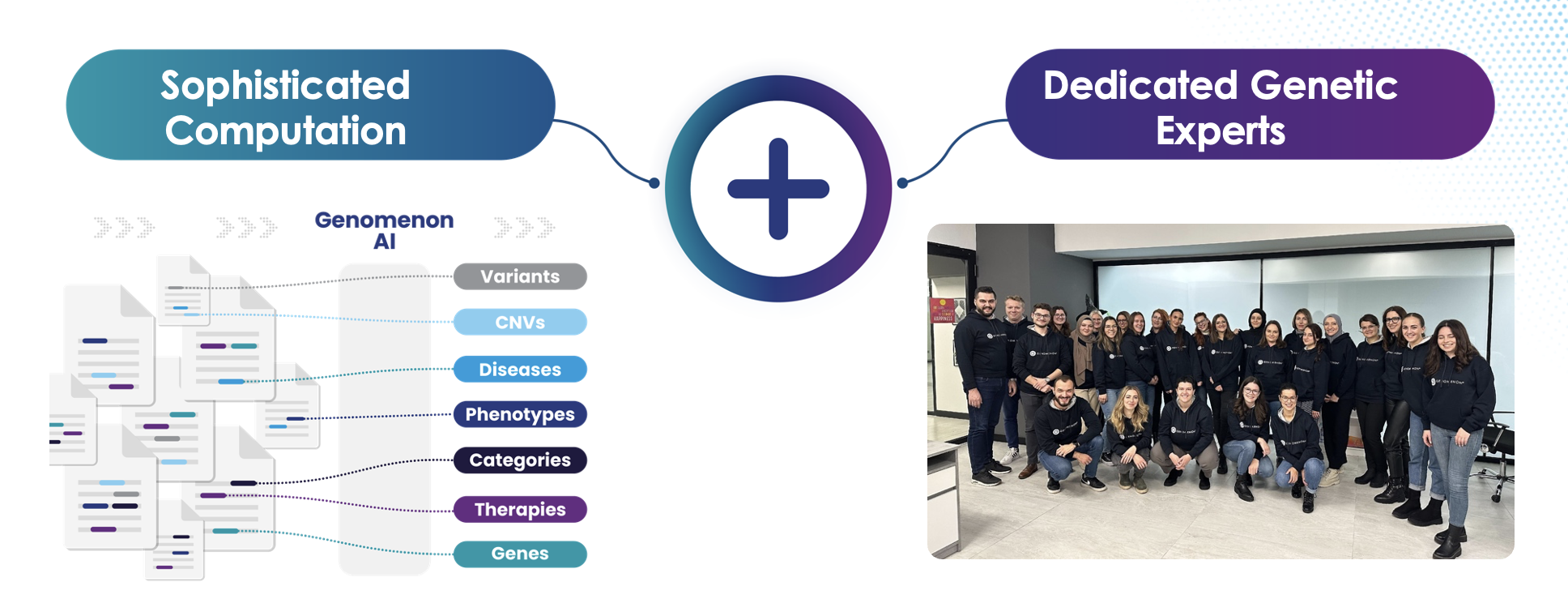
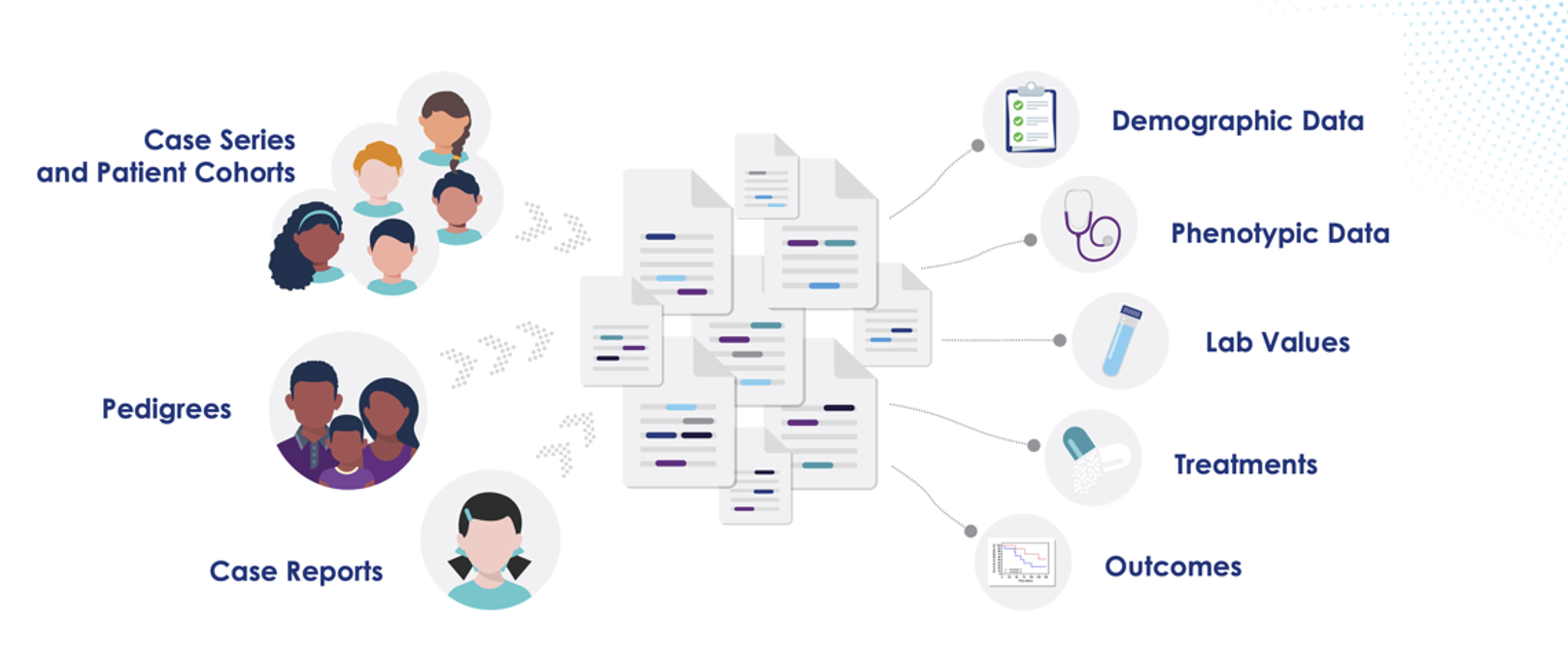
- Real-World Genomic Data is the New Frontier in Drug Development Genomenon’s approach combines expert human curation with advanced AI-powered tools to capture granular data from millions of full-text scientific articles. This data, structured in a proprietary graph database (G3), encompasses patient demographics, phenotypes, genetic variants, treatments, and outcomes.
- G3: A Scalable Framework for Evidence Discovery Jonathan Eads demonstrated how G3, Genomenon’s genomic graph database, enables structured extraction of clinical data to build powerful patient cohorts. Through a case study on colorectal cancer and immunotherapy, he illustrated how G3 identified a surprising responder subgroup: men under 50 with right-sided colon cancer, microsatellite stable status, and high tumor mutational burden. This finding contrasts with existing clinical assumptions, showcasing how literature-derived evidence can reveal underrecognized trends.
- Rare Disease Curation Can Shift the Diagnostic Landscape Dr. Sarah Chang shared a powerful example of real-world impact through UCB's work with Genomenon to resolve variants associated with thymidine kinase 2 deficiency (TK2D). By curating and submitting 95 TK2 variants to ClinVar, including many previously unrepresented in public databases, they improved the rate of variant resolution and diagnostic accuracy. Notably, one VUS (variant of uncertain significance) was reclassified as likely pathogenic thanks to a newly published case study and Genomenon’s curation.
- Mastermind Helps Reduce the Diagnostic Odyssey Through Mastermind, Genomenon’s flagship Genomic Intelligence Platform, users gain access to indexed, literature-backed variant data that would otherwise take years to manually collect. For rare disease applications, this is particularly transformative, helping patients avoid years of uncertainty and misdiagnosis.
- Real-World Evidence Can Scale, Inform, and Guide Precision Therapeutics The webinar emphasized that combining natural language processing with human curation offers a reproducible and high-fidelity method for variant and patient landscape analysis. With G3 and Mastermind, researchers and pharma teams can explore disease prevalence, identify biomarkers, and inform trial design with a level of precision previously impossible.
🔍 Audience Pulse Check: What the Polls Revealed
The live poll results during the webinar offered valuable insight into how professionals today interact with real-world evidence (RWE) derived from the literature - and where challenges remain.
When asked which data types are most difficult to extract at scale, phenotypic information stood out. This aligns with what many in genomics and drug development already know: while phenotypic details are among the most clinically useful, they’re often scattered, inconsistently reported, and hidden deep within the text of full publications. Unsurprisingly, this makes them especially difficult to systematize and apply at scale without powerful extraction tools.
When it comes to barriers to using literature-based RWE, trust in AI-powered extraction methods and limited access to advanced tools were both highlighted as significant concerns. This reveals a dual tension: while the potential value of literature is widely acknowledged, there’s hesitancy around the reliability of AI solutions and uncertainty about whether tools exist that can make this work scalable and accurate enough for high-stakes decision-making.
These poll responses highlight not just the current challenges in literature-based RWE, but also the readiness of the field to embrace tools that bridge the gap between potential and practice - particularly when they are transparent, scientifically rigorous, and built to handle the scale and nuance of real-world genomic data.
We’re incredibly grateful to those who took a moment to respond to our live poll during the session. At Genomenon, we see these insights not just as data points, but as the pulse of the audience - helping us understand the real challenges and priorities faced by those working on the frontlines of precision medicine. Your feedback directly shapes the conversations we lead, the tools we build, and the future topics we explore. Thank you for being part of the dialogue.
Final Thoughts
If you couldn’t attend the webinar, you missed a thorough walkthrough of how Genomenon is redefining what real-world evidence can look like in clinical and pharma contexts. From AI-powered graph databases to real-life VUS reclassification, the session spotlighted the tools and strategies that are making personalized medicine more efficient, equitable, and precise.
We invite you to watch the full webinar recording and reach out with questions or collaboration ideas. The future of genomic-driven precision medicine is here - and Genomenon is helping lead the way.







