In the test described below, the Mastermind Genomic Search Engine was found to have several meaningful advantages over Google Scholar as a tool for searching genomic evidence, reducing the amount of time required to curate a variant and resulting in fewer missed articles containing evidence for the variant. Users who searched Mastermind as part of this experiment felt more confident that they had found all of the relevant literature to form the most accurate diagnoses and treatment plans.
For over a quarter of the variants tested, Google Scholar was missing true positive articles from its search results. Additionally, for over half of the variants tested, Google Scholar had a significant number of false positive articles in its results which needed to be read by variant scientists to exclude.
Using Mastermind as a primary source for variant search, supplemented by Google Scholar, can ensure the most comprehensive and accurate results.
The Challenge of Variant Interpretation with Google Scholar
Variant interpretation requires extensive literature curation. First you must Identify the articles containing the most accurate and up-to-date information. Then you must draw clinically relevant determinations about the significance of a given variant. This process can require hours of painstaking manual review to draw well-informed conclusions, including these three steps:
- Finding articles mentioning a variant of interest
- Sifting through each article to identify the most high-yield resources
- Reading the prioritized references to find the critical descriptions of the data
Since more than 95% of variant citations are not found in the title or abstract of a manuscript, variant scientists rely on Google Scholar (GS) as a search tool to identify these references. But I have found in my clinical work that GS chronically identifies many off-target results:
- Results frequently contain false positive mentions
- Results are not organized in a clinically relevant order, but are rather prioritized based on text-frequency parameters
- Searchers must manually hunt through the full text of each paper to identify the mentions for confirmation
Moreover, once a relevant reference has been identified in GS search results, the searcher must leave the page and go out to the publication source to find, download, and sift through the entire article to confirm that the identification of the variant is accurate and not a false positive.
Mastermind Addresses Google Scholar Shortcomings
I encountered these challenges in my clinical and research practice when using Google Scholar, and I thought, “Why can’t this data come to me?” instead of needing to go out to the data each time I searched for a variant. Mastermind is a tool designed to do just that.
The Mastermind Genomic Search Engine uses automated techniques to index the full text of millions of highly prioritized articles and provides search results that eliminate the need to scour the references manually.
Mastermind Automatically:
- Identifies articles containing a given gene and variant
- Prioritizes the articles most likely to contain meaningful content
- Clearly presents the evidence for accurate and meaningful gene and variant matches
- Below are some screenshots of an example variant search in the Mastermind Genomic Search Engine:
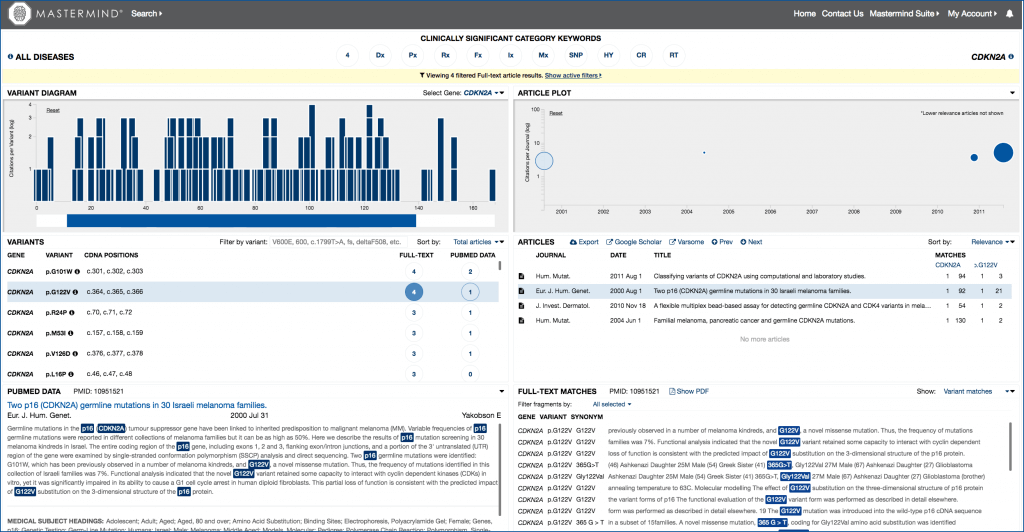
Fig. 1: Search results for CDKN2A and p.G122V in the Mastermind user interface

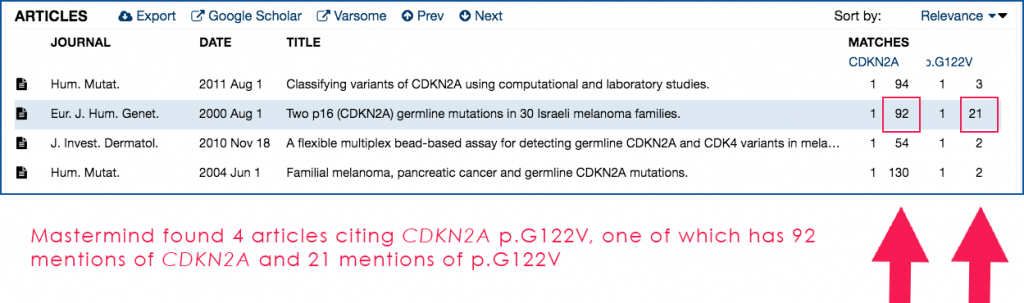
Fig. 2: Close-up of the Mastermind Article Results pane from Fig. 1
Fig. 3: Close-up of the Mastermind FULL-TEXT MATCHES pane from Fig. 1

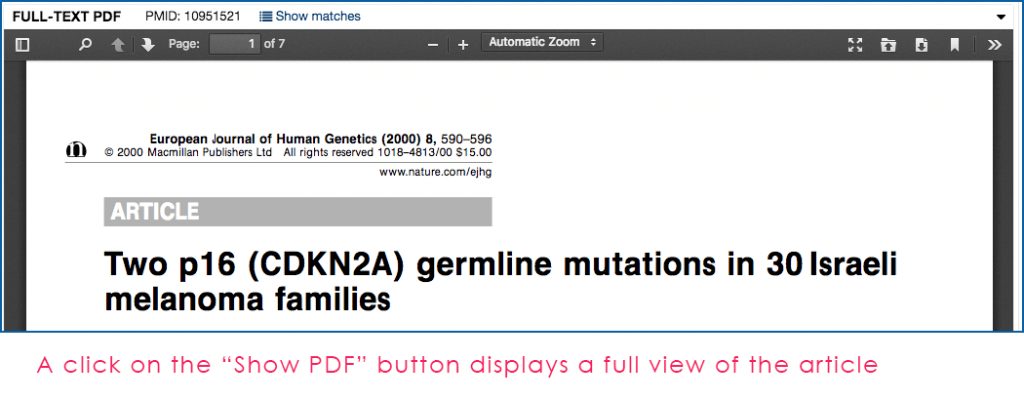
Fig. 4: Example of the Mastermind Full-Text PDF viewer for an article found in the search above
All of this information is organized by disease-gene-variant relationships in the Mastermind database, and can be queried using the web-based user interface.
Mastermind prioritizes and displays results that cut the time it takes to perform the literature curation required for variant interpretation. A common question we are asked is how the accuracy of Mastermind search results compares with GS. Namely, does Mastermind find the same articles as GS, and is it able to reduce the number of off-target results that can sometimes be seen when performing GS searches? This gets at the sensitivity and specificity of Mastermind search results as compared with a commonly used tool such as GS. Read on for the results of our test.
Search Result Accuracy Test: Google Scholar vs. Mastermind
To understand how Mastermind results compare with those of Google Scholar, we asked several of our users for a random selection of variants encountered in their clinical workflow that they had previously assessed using GS to identify articles. The same users then manually reviewed those articles to determine whether they in fact contained the variant or were false positive results. We then asked them to determine whether the same articles were found using a Mastermind search. The results were analyzed to determine Mastermind sensitivity and specificity, and to further determine whether GS missed any articles that were likely to contribute to a more accurate interpretation of the clinical significance of the variant.
In total, we received 192 variant search examples from users. Each variant was searched in GS using a multi-parameter search to ensure that both cDNA and protein level descriptions of the variant would be found (as typifies most GS search practices). An example search is presented below, given as the GS search criteria with the equivalent Mastermind search criteria.
Google Scholar search — “cdkn2a” ( “cdkn2a” ( “p.Gly122Val” | “Gly122Val” | “G122V” | “p.G122V” | “c.365G>T” | “365G>T” | “G365T” | “365G/T” ) )
Mastermind search — Gene: CDKN2A Variant: G122V
The articles as identified by PMID resulting from both the Mastermind and GS searches were enumerated and then manually reviewed and scored as being true positive (TP, article containing the variant) or false positive (FP, article containing something other than the variant or otherwise matching the variant but belonging to a different gene).
Results
Overall for the 192 variants tested, Mastermind performed very favorably compared to GS for the True Positive rate, and especially for the False Positive Rate.
Fig. 5 A table of the True Positive findings based on manual review. Mastermind found more PMIDs than GS in 25.5% of these cases, compared with only 9.4% of cases where GS found more PMIDs than Mastermind.
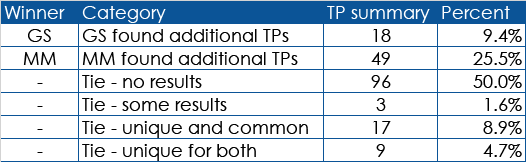
Fig. 6 A table of the False Positive findings based on manual review. Google Scholar returned false positive PMIDs in 67.7% of cases (52.1% + 15.6%). Mastermind returned far fewer False Positive PMIDs, and on a per case basis returned False Positives in only 16.6% of cases (1.0% + 15.6%).

Discussion: Google Scholar vs. Mastermind
Mastermind clearly out-performed Google Scholar as a genomic search engine when the test data for the 192 variants was summarized:
- Mastermind performed identically to Google Scholar in terms of sensitivity
- Mastermind outperformed Google Scholar in terms of specificity
- Mastermind finds as many articles as Google Scholar for any given variant
- Mastermind performs better than Google Scholar in reducing the number of false positive results
The improved performance of Mastermind compared to GS in terms of the accuracy of results is only one consideration. Other benefits of Mastermind which increase the efficiency of the variant interpretation process include:
- The ease of performing Mastermind searches without the need to devise Boolean-style nomenclature searches
- The prioritization of the search results according to any one of a number of clinically relevant categories
- The ease with which the gene, variant, and clinically relevant key term match contexts appear in the user interface at the user’s fingertips
- The ability of Mastermind to be automated using custom-designed APIs to further augment workflow efficiency.
In summary, using Mastermind as a primary source for variant search, supplemented by Google Scholar, can ensure the most comprehensive and accurate results.
Learn more and try Mastermind for yourself. Register for the Free version and get an upgrade to Mastermind Professional for your first 30 days.